Membrane Structure - Bio 5068
advertisement
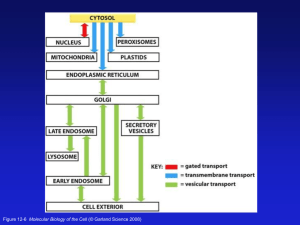
The Structure of Biological Membranes Thursday 8/28 2014 Mike Mueckler mmueckler@wustl.edu Functions of Cellular Membranes 1. Plasma membrane acts as a selectively permeable barrier to the environment • Uptake of nutrients • Waste disposal • Maintains intracellular ionic milieu 2. Plasma membrane facilitates communication • With the environment • With other cells 3. Intracellular membranes allow compartmentalization and separation of different chemical reaction pathways • Increased efficiency through proximity • Prevent futile cycling through separation • Protein secretion Composition of Animal Cell Membranes • Hydrated, proteinaceous lipid bilayers • By weight: 20% water, 80% solids • Solids: Lipid ~90% Protein Carbohydrate (~10%) • Phospholipids responsible for basic membrane bilayer structure and physical properties • Membranes are 2-dimensional fluids into which proteins are dissolved or embedded The Most Common Class of Phospholipid is Formed from a Gycerol-3-P Backbone Saturated Fatty Acid •Palmitate and stearate most common •14-26 carbons •Even # of carbons Unsaturated Fatty Acid Structure of Phosphoglycerides All Membrane Lipids are Amphipathic Figure 10-2 Molecular Biology of the Cell (© Garland Science 2008) Phosphoglycerides are Classified by their Head Groups Phosphatidylethanolamine Phosphatidylcholine Phosphatidylserine Ether Bond at C1 Phosphatidylinositol PS and PI bear a net negative charge at neutral pH Sphingolipids are the Second Major Class of Phospholipid in Animal Cells Sphingosine Ceramides contain sugar moities in ether linkage to sphingosine Glycolipids are Abundant in Brain Cells Figure 10-18 Molecular Biology of the Cell (© Garland Science 2008) Membranes are formed by the tail to tail association of two lipid leaflets Exoplasmic Leaflet Cytoplasmic Leaflet Bilayers are Thermodynamically Stable Structures Formed from Amphathic Lipids (Hydrophobic) (Hydrophilic) Figure 10-11a Molecular Biology of the Cell (© Garland Science 2008) Lipid Bilayer Formation is Driven by the Hydrophobic Effect • HE causes hydrophobic surfaces such as fatty acyl chains to aggregate in water • Water molecules squeeze hydrophobic molecules into as compact a surface area as possible in order to the minimize the free energy state (G) of the system by maximizing the entropy (S) or degree of disorder of the water molecules • DG = DH TDS Lipid Bilayer Formation is a Spontaneous Process Gently mix PL and water Vigorous mixing Figure 1-12 Molecular Biology of the Cell, Fifth Edition (© Garland Science 2008) The Formation of Cell-Like Spherical Water-Filled Bilayers is Energetically Favorable Figure 10-8 Molecular Biology of the Cell (© Garland Science 2008) Phospholipids are Mesomorphic Figure 10-7 Molecular Biology of the Cell (© Garland Science 2008) PhosphoLipid Movements within Bilayers (µM/sec) (1012-1013/sec) (108-109/sec) Figure 10-11b Molecular Biology of the Cell (© Garland Science 2008) Pure Phospholipid Bilayers Undergo Phase Transition Gauche Isomer Formation All-Trans Below Lipid Phase Transition Temp Above Lipid Phase Transition Temp Phosphoglyceride Biosynthesis Occurs at the Cytoplasmic Face of the ER Figure 12-57 Molecular Biology of the Cell (© Garland Science 2008) A “Scramblase” Enzyme Catalyzes Symmetric Growth of Both Leaflets in the ER Figure 12-58 Molecular Biology of the Cell (© Garland Science 2008) The Two Plasma Membrane Leaflets Possess Different Lipid Compositions Enriched in PC, SM, Glycolipids Enriched in PE, PS, PI Figure 10-16 Molecular Biology of the Cell (© Garland Science 2008) A “Flippase” Enzyme promotes Lipid Asymmetry in the Plasma Membrane Figure 12-58 Molecular Biology of the Cell (© Garland Science 2008) Membrane Proteins May Selectively Interact with Specific Lipids (Lipid Annulus or Halo) Lipid Asymmetry Also Exists in the Plane of the Bilayer Phospholipids are Involved in Signal Transduction 1. Activation of Lipid Kinases PI-4,5P Figure 10-17a Molecular Biology of the Cell (© Garland Science 2008) PI-3,4,5P 2. Phospholipases Produce Signaling Molecules via the Degradation of Phospholipids Phospholipase C Activation Produces Two Intracellular Signaling Molecules Diacylglycerol PI-3,4,5P I-3,4,5P Figure 10-17b Molecular Biology of the Cell (© Garland Science 2008) Amphipathic Lipids Containing Rigid Planar Rings Are Important Components of Biological Membranes Flexible Hydrocarbon Tail Hydrophilic End How Cholesterol Integrates into a Phospholipid Bilayer C12 Figure 10-5 Molecular Biology of the Cell (© Garland Science 2008) Cholesterol Biosynthesis Occurs in the Cytosol and at the ER Membrane Through Isoprenoid Intermediates (Rate-Limiting Step in ER) Lipid Rafts are Microdomains Enriched in Cholesterol and SM that may be Involved in Cell Signaling Processes Insoluble in Triton X-100 Figure 10-14b Molecular Biology of the Cell (© Garland Science 2008) Electron Force Microscopic Visualization of Lipid Rafts in Artificial Bilayers Yellow spikes are GPI-anchored protein Figure 10-14a Molecular Biology of the Cell (© Garland Science 2008) 3 Ways in which Lipids May be Transferred Between Different Intracellular Compartments Vesicle Fusion Direct Protein-Mediated Soluble Lipid Transfer Binding Proteins The 3 Basic Categories of Membrane Protein GPI Anchor Single-Pass Multi-pass b-Strands Transmembrane Helix Linker Domain Fatty acyl anchor Integral Figure 10-19 Molecular Biology of the Cell (© Garland Science 2008) LipidAnchored Peripheral (can also interact via PL headgroups) 3 Types of Lipid Anchors Figure 10-20 Molecular Biology of the Cell (© Garland Science 2008) Membrane Domains are “Inside-Out” Right-Side Out Soluble Protein Figure 3-5 Molecular Biology of the Cell (© Garland Science 2008) Functional Characterization of Integral Membrane Proteins Requires Solubilization and Subsequent Reconstitution into a Lipid Bilayer Figure 10-31 Molecular Biology of the Cell (© Garland Science 2008) Detergents are Critical for the Study of Integral Membrane Proteins (Used to denature proteins) (Used to purify Integral Membrane Proteins) Detergents Exist in Two Different States in Solution (Critical Micelle Concentration) Figure 10-29b Molecular Biology of the Cell (© Garland Science 2008) Detergent Solubilization of Membrane Proteins Desirable for Purification of Integral Membrane Proteins Beta Sheet Secondary Structure Polypeptide backbone is maximally extended (rise of 3.3 Å/residue) H-Bond Anti-parallel Strands Side chains alternately extend into opposite sides of the sheet b-Barrel Structure of the OmpX Porin Protein in the Outer Membrane of E. coli Aromatic Side Chain anchors Alternating Aliphatic Side Chains Transmembrane a-Helices H-Bond 1. Right-handed 2. Stability in bilayer results from maximum hydrogen bonding of peptide backbone 3. Usually > 20 residues in length (rise of 1.5 Å/residue) 4. Exhibit various degrees of tilt with respect to the membrane and can bend due to helix breaking residues 5. Mostly hydrophobic side-chains in single-pass proteins 6. Multi-pass proteins can possess hydrophobic, polar, or amphipathic transmembrane helices Transmembrane Domains Can Often be Accurately Identified by Hydrophobicity Analysis Figure 10-22b Molecular Biology of the Cell (© Garland Science 2008) Bacteriorhodopsin of Halobacterium Figure 10-32 Molecular Biology of the Cell (© Garland Science 2008) Structure of Bacteriorhodopsin Figure 10-33 Molecular Biology of the Cell (© Garland Science 2008) Structure of the Glycophorin A Homodimer Coiled-Coil Domains in Van Der Waals Contact Arg and Lys Side Chain Anchors 23 residue transmembrane helices Structure of the Photosynthetic Reaction Center of Rhodopseudomonas Viridis Figure 10-34 Molecular Biology of the Cell (© Garland Science 2008) 4 Ways that Protein Mobility is Restricted in Biological Membranes Intramembrane ProteinProtein Interactions Interaction with the cytoskeleton Interaction with the extracellular maxtrix Intercellular ProteinProtein Interactions Figure 10-39 Molecular Biology of the Cell (© Garland Science 2008)