Lecture11 - Department of Biological Sciences
advertisement
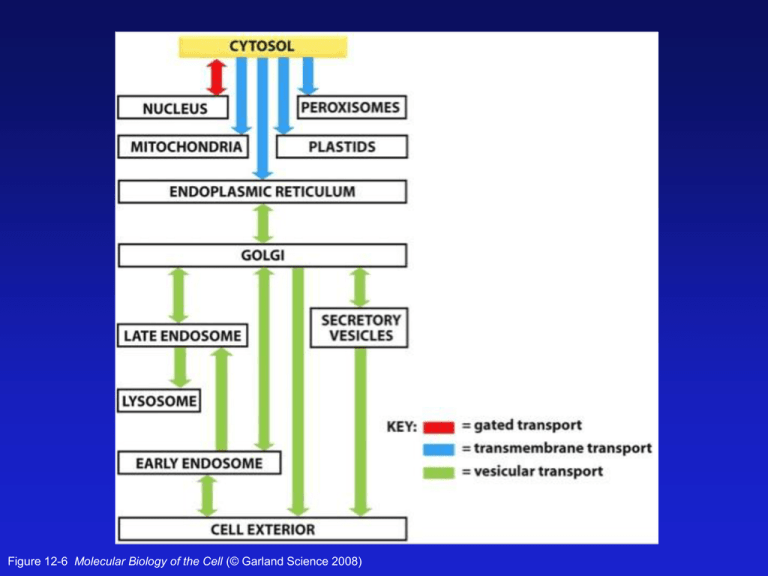
Figure 12-6 Molecular Biology of the Cell (© Garland Science 2008) Figure 12-21a Molecular Biology of the Cell (© Garland Science 2008) Some Proteins made in the mito. The Mitochondrial Import Sequence is an amphipathic alpha helix Figure 12-22 Molecular Biology of the Cell (© Garland Science 2008) TOM = cytosolic proteins to the intermembrane space TIM = cytosolic proteins to the matrix and inner membrane OXA = mitochondrially produced proteins to the intermembrane space Figure 12-23 Molecular Biology of the Cell (© Garland Science 2008) An in vitro experiment to determine how mitochondrial transport occurs Figure 12-24 Molecular Biology of the Cell (© Garland Science 2008) Cytosolic chaperones like Hsp70 keep proteins Unfolded until they are fed into the mitochondria And they contribute to the import process. Figure 12-25 Molecular Biology of the Cell (© Garland Science 2008) The RTG Network: a signaling pathway from the mitochondria to the Nucleus. Mitochondrial retrograde signaling is a pathway of communication from mitochondria to the nucleus under normal and pathophysiological conditions. The best understood of such pathways is retrograde signaling in the budding yeast Saccharomyces cerevisiae. It involves multiple factors that sense and transmit mitochondrial signals to effect changes in nuclear gene expression; these changes lead to a reconfiguration of metabolism to accommodate cells to defects in mitochondria. Analysis of regulatory factors has provided us with a mechanistic view of regulation of retrograde signaling. Here we review advances in the yeast retrograde signaling pathway and highlight its regulatory factors and regulatory mechanisms, its physiological functions, and its connection to nutrient sensing, TOR signaling, and aging. Aging, the greatest disease of all! Like many young scientists with a novel idea, Kenyon encountered more skepticism than support in the early 1990s. Indeed, one fellow scientist, worried that she had gone “over the edge,” warned that if she continued to insist that aging was subject to genetic regulation, she would soon fall off the Earth altogether. But her world turned out to be round, not flat. And now firmly anchored as, if not exactly the “queen of aging research,” then certainly its ace, Kenyon commands no fewer than 386,000 entries on a standard Google search. The story is now well-known. One of Kenyon’s lab rotation students — Ramon Tabtiang — in one of his very first experiments, picked a needle out of the haystack that is the C. elegans genome. In short, he found a mutant gene, dubbed daf-2, that made worms live twice as long. http://www.ucsf.edu/science-cafe/conversations/kenyon/ Peroxisome Zellweger syndrome PXR1 is a receptor on the surface of peroxisomes Peroxisome -self assembling, 1 day lifespan -arose due to oxygenation -proteins translated in cytosol, PTS directs them to peroxisomes -PTS1 in matrix proteins is usually Ser-Lys-Leu at the C-terminus -Pex5 is a PTS1 receptor that directs cytoplasmic proteins to per. -also a PTS2 sequence -membrane PEX proteins have a different targeting sequence Peroxisomal Biogenesis -integral membrane PEX10/12/2 form a receptor/channel on per. Peroxisomal Biogenesis -tetramer -> dimer of Pex5 Ines Heiland and Ralf Erdmann Peroxisomal Biogenesis Pex11 is involved in peroxisome propagation Peroxisomal Biogenesis Ines Heiland and Ralf Erdmann Peroxisomal Function and Enzymes 50 different enzymes Lipid metabolism Hydrogen peroxide breakdown: catalase Lipid biosynthesis (cholesterol) Amine and bile acid synthesis Purine catabolism by urate oxidase Peroxisomal Function and Enzymes ß-oxidation of Very Long Chain Fatty Acids (VLCFA) provides the cell with a major source of metabolic energy (oxidation of fats mostly occurs in mitos) Saturated Unsaturated (“good”) Peroxisomal Function and Enzymes 50 different enzymes Lipid metabolism: ß-oxidation of Very Long Chain Fatty Acids (VLCFA) -provides the cell with a major source of metabolic energy Hydrogen peroxide breakdown: catalase Lipid bisynthesis (cholesterol) Amine and bile acid synthesis Purine catabolism by urate oxidase Figure 12-54 Molecular Biology of the Cell (© Garland Science 2008)