1.3 Cell Membrane 2016
advertisement

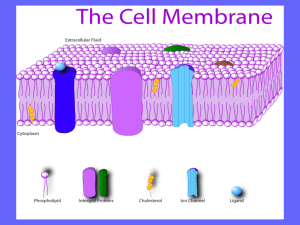
1.3 Membrane Structure Essential idea: The structure of biological membranes makes them fluid and dynamic. Above are models of a plasma membrane showing how it's fluidity allows lipid soluble molecules to move directly through the membrane. By Chris Paine https://bioknowledgy.weebly.com/ http://www.europhysicsnews.org/doc_journal/images/epn/hl/435/Sommer.jpg Understandings, Applications and Skills 1.3.U1 1.3.U2 1.3.U3 1.3.A1 1.3.S1 Statement Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. Membrane proteins are diverse in terms of structure, position in the membrane and function. Cholesterol is a component of animal cell membranes. Cholesterol in mammalian membranes reduces membrane fluidity and permeability to some solutes. Drawing of the fluid mosaic model. 1.3.S2 Analysis of evidence from electron microscopy that led to the proposal of the Davson-Danielli model. 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the Singer-Nicolson model. Guidance Amphipathic phospholipids have hydrophilic and hydrophobic properties. Drawings of the fluid mosaic model of membrane structure can be two dimensional rather than three dimensional. Individual phospholipid molecules should be shown using the symbol of a circle with two parallel lines attached. A range of membrane proteins should be shown including glycoproteins. 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. What happens when you put a drop of oil in water? http://www.flickr.com/photos/zorin-denu/5385963280/ 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. The Oil droplet stays together and makes a perfect circular shape. The oil molecules are Hydrophobic Oil Molecules are nonpolar and water molecules are polar. See 3.1.5 http://www.flickr.com/photos/zorin-denu/5385963280/ 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. Phospholipid molecules have a polar (charged) phosphate head and long non-polar lipid tails • The head is hydrophillic (attracted to water) • The tails are hydrophobic (repelled by water) When drawing a diagram of a phospholipid this is a good example which shows all the key features http://www.ib.bioninja.com.au/_Media/phospholipid_bilayer_med.jpeg http://upload.wikimedia.org/wikipedia/commons/3/39/Phospholipid_TvanBrussel.jpg 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. When put into water, an emergent property is that phospholipids will self-organise to keep their heads ‘wet’ and their tails ‘dry’ micelle liposome http://commons.wikimedia.org/wiki/File:Micelle_scheme-en.svg http://commons.wikimedia.org/wiki/File:Liposome_scheme-en.svg 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. In this 3D representation you can see that a phospholipid bilayer is one way that the tails can be removed from the water. Phospholipid molecules can flow past each other laterally but can’t move vertically http://commons.wikimedia.org/wiki/File:Phospholipids_aqueous_solution_structures.svg 1.3.U1 Phospholipids form bilayers in water due to the amphipathic properties of phospholipid molecules. But wait! there’s more! The plasma membrane is not just made of phospholipids http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.U2 Membrane proteins are diverse in terms of structure, position in the membrane and function. Proteins: Integral proteins are permanently embedded, many go all the way through and are polytopic (poly = many, topic = surface), integral proteins penetrating just one surface are monotopic. Peripheral proteins usually have a temporary association with the membrane, they can be monotopic or attach to the surface http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.U2 Membrane proteins are diverse in terms of structure, position in the membrane and function. Glycoproteins: Are proteins with an oligosaccaride (oligo = few, saccharide = sugar) chain attached. They are important for cell recognition by the immune system and as hormone receptors http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.U2 Membrane proteins are diverse in terms of structure, position in the membrane and function. Transport: Protein channels (facilitated) and protein pumps (active) Receptors: Peptide-based hormones (insulin, glucagon, etc.) Anchorage: Cytoskeleton attachments and extracellular matrix Cell recognition: MHC proteins and antigens Intercellular joinings: Tight junctions and plasmodesmata Enzymatic activity: Metabolic pathways (e.g. electron transport chain) http://www.ib.bioninja.com.au/standard-level/topic-2-cells/24-membranes.html 1.3.U3 Cholesterol is a component of animal cell membranes. Cholesterol: (It’s not all bad!) It makes the phospholipids pack more tightly and regulates the fluidity and flexibility of the membrane. Bad analogy: imagine a room full of people wearing fluffy jumpers (sweaters). It is crowded but they can slip past each other easily enough. Now sprinkle the crowd with people wearing Velcro™ suits… 1.3.U3 Cholesterol is a component of animal cell membranes. Cholesterol Hydroxyl group makes the head polar and hydrophilic - attracted to the phosphate heads on the periphery of the membrane. Carbon rings – it’s not classed as a fat or an oil, cholesterol is a steroid Non-polar (hydrophobic) tail –attracted to the hydrophobic tails of phospholipids in the centre of the membrane http://www.uic.edu/classes/bios/bios100/lectf03am/cholesterol.jpg http://www.cholesterol-and-health.com/images/Cholesterol_Structure.jpg 1.3.A1 Cholesterol in mammalian membranes reduces membrane fluidity and permeability to some solutes. Membrane fluidity The hydrophobic hydrocarbon tails usually behave as a liquid. Hydrophilic phosphate heads act more like a solid. Though it is difficult to determine whether the membrane is truly either a solid or liquid it can definitely be said to be fluid. It is important to regulate the degree of fluidity: • Membranes need to be fluid enough that the cell can move • Membranes need to be fluid enough that the required substances can move across the membrane • If too fluid however the membrane could not effectively restrict the movement of substances across itself http://www.nature.com/scitable/content/ne0000/ne0000/ne0000/ne0000/14668965/U2.cp5.3_membrane_f2.jpg 1.3.A1 Cholesterol in mammalian membranes reduces membrane fluidity and permeability to some solutes. Cholesterol’s role in membrane fluidity 1. The presence of cholesterol in the membrane restricts the movement of phospholipids and other molecules – this reduces membrane fluidity. 2. The presence of cholesterol disrupts the regular packing of the of the hydrocarbon tails of phospholipid molecules - this is increases the flexibility as it prevents the tails from crystallising and hence behaving like a solid. 3. Cholesterol also reduces the permeability to hydrophilic/water soluble molecules and ions such as sodium and hydrogen. http://www.nature.com/scitable/content/ne0000/ne0000/ne0000/ne0000/14668965/U2.cp5.3_membrane_f2.jpg 1.3.S1 Drawing of the fluid mosaic model. Use the tutorials to learn and review membrane structure http://www.bio.davidson.edu/people/macampbell/111/membswf/membranes.swf http://www.phschool.com/science/biology_place/biocoach/biomembrane1/regi ons.html https://www.wisc-online.com//LearningContent/ap1101/index.html 1.3.S1 Drawing of the fluid mosaic model. http://www.youtube.com/watch?v=w9VBHGNoFrY 1.3.S1 Drawing of the fluid mosaic model. Reminder of features that make good diagrams: • • • • • Good use of space • Clear strong lines Label lines are straight • Labels clearly written (Scale bar if appropriate) Lines touch the labeled structure No unnecessary shading or colouring http://www.ib.bioninja.com.au/_Media/phospholipid_bilayer_med.jpeg 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the SingerNicolson model. Our current model of the cell membrane is called the Singer-Nicholson fluid mosaic model Key features: • • • • • Phospholipid molecules form a bilayer - phospholipids are fluid and move laterally Peripheral proteins are bound to either the inner or outer surface of the membrane Integral proteins - permeate the surface of the membrane The membrane is a fluid mosaic of phospholipids and proteins Proteins can move laterally along membrane http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the SingerNicolson model. Our current model of the cell membrane is called the Singer-Nicholson fluid mosaic model There is strong evidence for this model: Biochemical techniques • Membrane proteins were found to be very varied in size and globular in shape • Such proteins would be unable to form continuous layers on the periphery of the membrane. • The membrane proteins had hydrophobic regions and therefore would embed in the membrane not layer the outside http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the SingerNicolson model. Our current model of the cell membrane is called the Singer-Nicholson fluid mosaic model There is strong evidence for this model: Fluorescent antibody tagging • red or green fluorescent markers attached to antibodies which would bind to membrane proteins • The membrane proteins of some cells were tagged with red markers and other cells with green markers. • Within 40 minutes the red and green markers were mixed throughout the membrane of the fused cell. • This showed that membrane proteins are free to move within the membrane rather than being • The cells were fused together. fixed in a peripheral layer. http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the SingerNicolson model. Our current model of the cell membrane is called the Singer-Nicholson fluid mosaic model This model was first proposed in by Singer-Nicolson in 1972 Before then Davson-Danielli model was widely accepted … http://commons.wikimedia.org/wiki/File:Cell_membrane_detailed_diagram_en.svg?uselang=en-gb 1.3.S2 Analysis of evidence from electron microscopy that led to the proposal of the Davson-Danielli model. The evidence: In high magnification electron micrographs membranes appeared as two dark parallel lines with a lighter coloured region in between. Proteins appear dark in electron micrographs and phospholipids appear light - possibly indicating proteins layers either side of a phospholipid core. Davson-Danielli Model The model: • A protein-lipid sandwich • Lipid bilayer composed of phospholipids (hydrophobic tails inside, hydrophilic heads outside) • Proteins coat outer surface • Proteins do not permeate the lipid bilayer Proteins Pore Phospholipids This explains: Despite being very thin membranes are an effective barrier to the movement of certain substances. http://www.cytochemistry.net/cell-biology/EMview.jpg http://upload.wikimedia.org/wikipedia/commons/6/69/Davson_danielli_miguelferig.jpg 1.3.S3 Analysis of the falsification of the Davson-Danielli model that led to the SingerNicolson model. Falsification of the Davson-Danielli model – freeze fracturing This technique involves rapid freezing of cells and then fracturing them. Interpreting the image: • The fracture occurs along lines of weakness, including the centre of membranes. • The fracture reveals an irregular rough surface inside the phospholipid bilayer • The globular structures were interpreted as transmembrane proteins. Conclusion: This is contrary to the Davson-Danielli model which only involves proteins coating the surface of the membrane. A new model is needed to explain the presence of as trans-membrane proteins. http://www.cytochemistry.net/cell-biology/ffimage.jpg Bibliography / Acknowledgments Jason de Nys