CHEM642-09 Powerpoint
advertisement

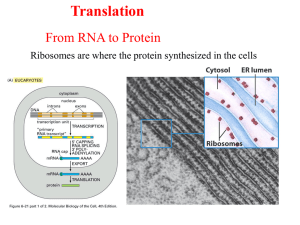
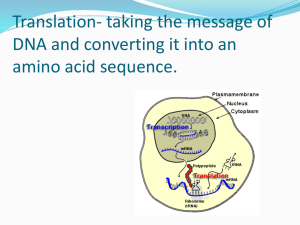
Translation From RNA to Protein Ribosomes are here the protein synthesized in the cells Steps of Translation MESSENGER RNA Polypeptide chains are specified by open reading frames Three possible reading frames of E. coli trp leader sequence An mRNA sequence is decoded in set of three nucleotides The standard one-letter abbreviation for each amino acid is presented below its three-letter abbreviation (see Panel 3–1, pp. 132–133, for the full name of each amino acid and its structure). By convention, codons are always written with the 5'- terminal nucleotide to the left. Note that most amino acids are represented by more than one codon, and that there are some regularities in the set of codons that specifies each amino acid. Codons for the same amino acid tend to contain the same nucleotides at the first and second positions, and vary at the third position. Three codons do not specify any amino acid but act as termination sites (stop codons), signaling the end of the protein- coding sequence. One codon— AUG—acts both as an initiation codon, signaling the start of a protein-coding message, and also as the codon that specifies methionine. Prokaryotic mRNAs have a ribosome-binding site that recruits the translational machinery Eukaryotic mRNAs are modified at their 5’ and 3’ ends to facilitate translation TRANSFER RNA tRNA molecules match amino acids to codons in mRNA Crick’s adaptor hypothesis A subset of modified nucleotides found in tRNA tRNAs share a common secondary structure that resembles a cloverleaf The anticodon is the sequence of three nucleotides that base-pairs with a codon in mRNA. The amino acid matching the codon/anticodon pair is attached at the 3' end of the tRNA. tRNAs contain some unusual bases, which are produced by chemical modification after the tRNA has been synthesized. For example, the bases denoted y and D are derived from uracil. (B and C) Views of the actual L-shaped molecule, based on x-ray diffraction analysis. Although a particular tRNA, that for the amino acid phenylalanine, is depicted, all other tRNAs have very similar structures. (D) The linear nucleotide sequence of the molecule, color-coded to match A, B, and C. tRNAs have an L-shaped 3 dimensional structures ATTACHMENT OF AMINO ACIDS TO tRNA tRNAs are charged by attachment of an AA to its 3’ terminal Aminoacyl-tRNA synthetases charge tRNAs in two steps Each aminoacyl-tRNA synthetase attaches a single amino acid to one or more tRNAs An mRNA sequence is decoded in set of three nucleotides The standard one-letter abbreviation for each amino acid is presented below its three-letter abbreviation (see Panel 3–1, pp. 132–133, for the full name of each amino acid and its structure). By convention, codons are always written with the 5'- terminal nucleotide to the left. Note that most amino acids are represented by more than one codon, and that there are some regularities in the set of codons that specifies each amino acid. Codons for the same amino acid tend to contain the same nucleotides at the first and second positions, and vary at the third position. Three codons do not specify any amino acid but act as termination sites (stop codons), signaling the end of the protein- coding sequence. One codon— AUG—acts both as an initiation codon, signaling the start of a protein-coding message, and also as the codon that specifies methionine. tRNA synthetases recognize unique structural features of cognate tRNAs tRNA elements required for aminoacy-tRNA synthetase recognition The recognition of a tRNA molecule by its aminoacyl-tRNA synthetase. For this tRNA (tRNAGln), specific nucleotides in both the anticodon (bottom) and the amino acid-accepting arm allow the correct tRNA to be recognized by the synthetase enzyme (blue). (Courtesy of Tom Steitz.) Aminoacyl-tRNA formation is very accurate Some aminoacyl-tRNA synthetases use an editing pocket to charge tRNAs with high accuracy Hydrolytic editing. (A) tRNA synthetases remove their own coupling errors through hydrolytic editing of incorrectly attached amino acids. As described in the text, the correct amino acid is rejected by the editing site. (B) The error-correction process performed by DNA polymerase shows some similarities; however, it differs so far as the removal process depends strongly on a mispairing with the template. The ribosome is unable to discriminate between correctly and incorrectly charged tRNAs THE RIBOSOME Prokaryotic RNAP and ribosomes at work on the same mRNA The ribosome is composed of a large and small subunit The large and small subunits undergo association and dissociation during each cycle of translation Each mRNA can be translated simultaneously by multiple ribosomes New amino acids are attached to the carboxyl terminus of the growing peptide chain Ribosomal RNAs are both structural and catalytic determinants of the ribosome Two views of ribosome The ribosome has three binding sites for tRNA Views of 3-D structure of the ribosome including three bound tRNAs Channels through the ribosome allow the mRNA and growing polypeptide to enter and/or exit the ribosome Interactions of tRNAs with mRNA The path of mRNA through the small ribosomal subunit The polypeptide exit tunnel The Nobel Prize in Chemistry 2009 "for studies of the structure and function of the ribosome" Venkatraman Ramakrishnan Thomas A. Steitz Ada E. Yonath 1/3 of the prize 1/3 of the prize 1/3 of the prize United Kingdom USA Israel MRC Laboratory of Molecular Biology Cambridge, United Kingdom Yale University New Haven, CT, USA; Howard Hughes Medical Institute Weizmann Institute of Science Rehovot, Israel b. 1952 (in Chidambaram, Tamil b. 1940 b. 1939 INITIATION OF TRANSLATION Prokaryotic mRNAs are initially recruited to the small subunit by base pairing to rRNA A specialized tRNA charged with a modified methionine binds directly to prokaryotic small subunit Three initiation factors direct the assembly of an initiation complex that contains mRNA and the initiator tRNA 14_Figure25a.jpg 14_Figure25b.jpg 14_Figure25c.jpg 14_Figure25d.jpg 14_Figure25e.jpg 14_Figure25f.jpg Eukaryotic ribosomes are recruited to the mRNA by the 5’ cap The start codon is found by scanning downstream from the 5’ end of the mRNA Translation initiation factors hold eukaryotic mRNAs in circles TRANSLATION ELONGATION Aminoacyl-tRNAs are delivered to the A site by elongation factor EF-Tu The ribosome uses multiple mechanisms to select against incorrect aminoacyl-tRNAs The ribosome is a ribozyme RNA surrounds the peptidyl transferase center of the large ribosomal subunit Proposed role for the 2’-OH of the P site tRNA in peptide bond formation Peptide bond formation and the elongation factor EF-G drive translocation of the tRNAs and the mRNA EF-G drive translocation by displacing the tRNA bound to the A site Structural comparison of EF-Tu-tRNA and EF-G EF-TU-GDP and EF-G-GDP must exchange GDP for GTP prior to participating in a new round of elongation A cycle of peptide bond formation consumes two GTPs and one ATP TERMINATION OF TRANSLATION Release factors terminate translation in response to stop codon Short regions of class I release factors recognize stop condons and trigger release of the peptide chain 3-D structures of RF1 bound to the ribosome Comparison of the structures of RFI to a tRNA GDP/GTP exchange and GTP hydrolysis control the release of the class II release factor The ribosome recycling factor (RRF) mimics a tRNA to stimulate the release of tRNA and mRNA from a terminated ribosome