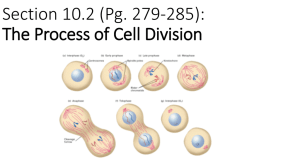
Chapter 6B
advertisement

Topics Chap. 6 Genes, Genomics, and Chromosomes (Part B) • Genomics: Genome-wide Analysis of Gene Structure and Expression • Structural Organization of Eukaryotic Chromosomes • Morphology and Functional Elements of Eukaryotic Chromosomes Goals • Learn about computer-based methods for analyzing sequence data. • Learn how DNA and proteins are packaged in chromatin. • Learn the large-scale structure organization of chromosomes. • Learn the functional elements required for chromosome replication and segregation. RxFISH-painted human chromosomes. Mining Sequence Data: BLAST Searches An enormous amount of DNA sequence information is available from genome sequencing and sequencing of cloned genes. This data is stored in data banks such as GenBank at the NIH in Bethesda, MD and the EMBL Sequence Data Base at the European Molecular Biology Laboratory in Heidelberg, Germany. Scientists working in the area of bioinformatics use this data to find genes, analyze their properties, and determine phylogenetic relationships between organisms and proteins. A common procedure in which this data is used is the BLAST search (basic local alignment search tool) which is used to compare protein and DNA sequences. An example BLAST search alignment is shown for the human neurofibromatosis 1 (NF1) gene in Fig. 6.25. The alignment shows NF1 is related to the S. cerevisiae Ira GTPase-activating protein (GAP) and suggests the disease is caused by aberrant signal transduction. Computer programs similar to BLAST are used to identify protein sequence motifs (e.g., zinc fingers) in unknown proteins. The identification of structure regions with known function sheds light on overall protein function and helps guide experimental analysis of unknown proteins and genes. Sequence Comparisons Establish Evolutionary Relationships Among Proteins BLAST search analysis can identify the members of a protein family originating from gene duplication and speciation mutations. As illustrated for the a- and ß-tubulin protein family in Fig. 6.26, an early gene duplication event created the paralogous aand ß-tubulin genes. Later speciation mutations lead to evolution of the orthologous members of the a- and ß-tubulin subfamilies. Orthologous proteins are most likely to share the same function. x Genome Size vs Complexity Genome sequencing has revealed that the morphological complexity of an organism is not strongly correlated with the size of its genome (Fig. 6.27). Alternative splicing of RNAs and posttranslational modification of proteins are thought to greatly increase the complexity of the proteins encoded by the genomes of higher organisms. In addition, the relative number of cells formed in a tissue such as the cerebral cortex can be important in increasing complexity (e.g., mice vs humans). Genes can be identified within the sequenced genomes of simple organisms such as yeast and bacteria by searching for open reading frames (ORFS). ORFs are long stretches of triplet codons lacking stop codons. Gene annotation (assignment of likely function) is based on knowledge from biochemical studies and/or alignments with known sequences. In complex organisms such as humans whose genes typically contain introns, more sophisticated algorithms that ID intron splice sites and compare cDNA and other sequence information to genomic DNA sequences must be applied to locate and annotate genes. Using such methods ~25,000 genes have been identified in humans. However, conclusive evidence for synthesis of protein or RNA products is lacking for ~10,000 genes. Extended and Condensed Chromatin Human diploid cells contain about 2 meters of DNA. To fit within nuclei, DNA must be condensed by ~105-fold. DNA exists in cells as a nucleoprotein complex known as chromatin. During interphase when cells are not dividing, chromatin is relatively uncondensed compared to its state in metaphase chromosomes. When released from nuclei with low salt buffer, chromatin displays an extended "beads-on-a-string" morphology, where each bead is a nucleosome (Fig. 6.28). When released in physiological salt concentrations, more condensed fibers of 30 nm diameter are observed. In general, extended chromatin can be transcribed, whereas condensed forms cannot. Structure of Nucleosomes Nucleosomes consist of 147 bp of DNA wrapped in almost two turns around the outside of an octamer of histone proteins (Fig. 6.29). In most nucleosomes, the octamer has a stoichiometry of H2A2H2B2H32H42. Histones are the most abundant DNA-binding proteins in eukaryotic cells. The sequences of the 4 histones that make up the octamer are highly conserved across all organisms, indicating their functions were optimized early in evolution. Histones have a large number of basic amino acids and bind to DNA mostly by salt-bridge interactions to phosphates in the DNA backbone. Another histone, H1, binds to the linker DNA between nucleosomes. Linker DNA is 10-90 bp in length depending upon the organism. Structure of 30-nm Chromatin Fibers In 30-nm fibers, nucleosomes bind to one another in a double helical arrangement (Fig. 6.30). Histone H1 molecules bind to linker DNA between nucleosomes and help stabilize the 30-nm fiber. The stability of 30-nm fibers is modulated by posttranslational modification of the tails of histones in the octamers (H4 in particular). Histone Tails and Chromatin Condensation The N- and C-terminal tails of histones project out from the nucleosome core (Fig. 6.31a). They also contain numerous residues that can be modified by acetylation, methylation, etc. (Fig. 6.31b). Acetylation of lysine side-chains by histone acetylases (HATs) neutralizes positive charge and promotes decondensation of 30-nm fibers. Methylation, on the other hand, blocks lysine acetylation, maintains positive charge, and promotes 30-nm fiber condensation. Studies have shown that chromatin condensation is not controlled simply by the net acetylation state of histones. Rather, the sites where acetylation and other modifications occur also are important. The combinations of modifications that specify condensation/decondensation are referred to as the "histone code". Interphase Chromatin Interphase chromatin exists in two different condensation states (Fig. 6.33a). Heterochromatin is a condensed form that has a condensation state similar to chromatin found in metaphase chromosomes. Euchromatin is considerably less condensed. Heterochromatin typically is found at centromere and telomere regions, which remain relatively condensed during interphase. The inactivated copy of the Xchromosome (Barr body) that occurs in cells in females also occurs as heterochromatin. In contrast, most transcribed genes are located in regions of euchromatin. Common modifications occurring in histone H3 in heteroand euchromatin are illustrated in Fig. 6.33b. Formation of Heterochromatin The trimethylation of histone H3 at lysine 9 (H3K9Me3) plays an important role in promoting chromatin condensation to heterochromatin (Fig. 6.34a). Trimethylated sites are bound by heterochromatin protein 1 (HP1) which self-associates and oligomerizes resulting in heterochromatin. Heterochromatin condensation is thought to spread laterally between “boundary elements” that mark the ends of transcriptionally active euchromatin (Fig. 6.34b). Recruitment of the H3K9 histone methyl transferase (HMT) to HP1 sites promotes heterochromatin spreading by catalyzing H3 methylation. Structure of Interphase Chromosomes FISH analysis performed with fluorescent probes that bind to sequential sequence sites along DNA supports a looped structure for interphase chromosomes (Fig. 6.35). Loops range in size from 1 to 4 million base pairs in mammalian interphase cells. The bases of the loops are located near the center of the chromosome at scaffold-associated regions (SARs), and matrixattachment regions (MARs). The DNA fibers at the base of the loops are held together by structural maintenance of chromosome (SMC) proteins (Fig. 6.36c) and other non-histone proteins. Transcription units containing expressed genes are located in uncondensed loop regions, away from the more condensed center of the chromosome. Interphase Chromosome Territories In situ hybridization of interphase nuclei with chromosomespecific fluorescently-labeled probes indicates that chromosomes reside within restricted regions of the nucleus rather than appearing throughout the nucleus (Fig. 6.37). Interestingly, the precise positions of chromosomes are not reproducible between cells. Structure of Metaphase Chromosomes In metaphase chromosomes, the number of loops of chromatin is increased and the lengths of the loops are decreased compared to what occurs in interphase chromosomes. In addition, more folded structures called chromonema fibers and higher order structures occur in prophase and metaphase chromatids (Fig. 6.38). Microscopic Structure of Metaphase Chromosomes Because interphase chromosomes are not easily visualized by microscopy techniques, chromosome morphology has been studied mostly using metaphase chromosomes. Metaphase chromosomes are duplicated structures formed after DNA replication is complete. They contain two sister chromatids joined at a structure called the centromere (Fig. 6.39). The ends of chromatids are called telomeres. Centromeres are required for chromatid separation late in mitosis. Telomeres are important in preventing chromosome shortening during replication. The number, sizes, and shapes of metaphase chromosomes constitute the karyotype, which is distinctive for each species. Chromosome Banding Patterns A number of dyes, such as Giemsa reagent, selectively stain different regions of chromosomes forming distinctive bands. For Giemsa reagent, banding is affected by G + C content. Banding patterns are very important in chromosome ID and in looking for chromosomal abnormalities and mapping the locations of genes. The most detailed staining is achieved via multicolor FISH chromosome painting. In this technique, staining is performed using a mixture of DNA probes coupled to several fluorescent dyes (See Slide 1). In Fig. 6.40 below, FISH staining patterns have been converted to false-color images to visualize chromosomes. Standard terminology is used for naming band and gene locations in chromosomes. The short arm is designated "p", and the long arm "q". Arms are further divided into major sections and subsections that are numbered consecutively out from the centromere. Detection of Translocations The analysis of chromosome banding patterns is used to detect anomalies such as truncations and translocations associated with certain genetic disorders and cancers. In chronic myelogenous leukemia, leukemic cells contain a shortened chromosome 22 and a longer chromosome 9 resulting from a translocation event in the q arms of these two chromosomes (Fig. 6.41). The shortened chromosome is distinctive and is referred to as the "Philadelphia chromosome". Multicolor FISH staining (right) is useful in identification of such chromosomes in a chromosome spread. Evolution of Human Chromosomes Through the determination of locations of common chromosomal segments in modern primate chromosomes, investigators have calculated the most likely karyotype of the common ancestor of all primates (Fig. 6.42c). In addition, they have proposed a model for how the human karyotype evolved from that ancestor. Major events in the evolution of the human karyotype include 1) formation of chromosome 2 by fusion of ancestral chromosomes 9 and 11, 2) formation of chromosomes 14 and 15 by breakage of ancestral chromosome 5, and 3) formation of chromosomes 12 and 22 by translocations between ancestral chromosomes 14 and 21. In other cases (e.g., chromosome 1), no significant rearrangements have occurred over time. ID of Functional Chromosomal Elements (I) Studies with yeast have demonstrated that all chromosomes must contain 3 functional elements to replicate and segregate correctly: 1) replication origins, 2) a centromere, and 3) telomeres. Yeast replication origins were identified in plasmid cloning studies. Only yeast plasmids containing a copy of a sequence referred to as the autonomously replicating sequence (ARS) could be transfected into yeast cells (Fig. 6.44a). The haploid S. cerevisiae genome contains many ARSs distributed among its 16 chromosomes. ID of Functional Chromosomal Elements (II) While only ARSs are needed for plasmid replication, an additional sequence identified by cloning procedures was found to be required for efficient segregation of plasmids to yeast daughter cells (Fig. 6.44b). This DNA proved to contain chromosomal centromere sequences (CEN sequences). Yeast CEN sequences are relatively simple (Fig. 6.45, not covered). In humans, they consist of 2-4 x 106 bp of simple sequence DNA composed of a 171 bp repeat unit. The human centromere sequence is bound by specialized nucleosomes containing a centromere-specific histone H3 variant (CENP-A). A large complex of non-histone proteins (the kinetochore) binds to centromeres and attaches them to microtubules of the mitotic spindle apparatus. ID of Functional Chromosomal Elements (III) Yeast transfection studies also showed that linearized plasmids containing ARS and CEN sequences could be maintained in cells only if telomere (TEL) sequences were attached at their ends (Fig. 6.44c). The function of TEL sequences in replication of chromosome ends is illustrated in the next two slides. Function of Telomeres A special mechanism is needed to complete the replication of DNA in DNA strands that have their 3’ ends located at the ends of chromosomes. DNA polymerases cannot complete synthesis of this region of DNA, and without synthesis, chromosomes become shortened with each round of replication (Fig. 6.46). Shortening results in the loss of binding sites for proteins that protect the ends of linear chromosomes from attack by exonucleases. As illustrations of the importance of telomere replication, knockout mice lacking the enzyme that synthesizes DNA at telomeres, telomerase, cannot produce viable offspring after six generations. In addition, telomerase often is switched on in cancer cells. Mechanism of Action of Telomerase Telomere sequences typically consist of tandemly repeating sequence units with a high G content in the strand that has its 3' end at the end of the chromosome. In humans and other vertebrates, the repeating sequence is TTAGGG. This sequence unit repeats over a few thousand base pairs in humans. The mechanism of replication of this DNA is illustrated in Fig. 6.47 for a protozoan species. Replication is carried out by the enzyme known as telomerase. Telomerase is a reverse transcriptase that carries its own internal RNA template which binds to the ssDNA at the chromosome 3’ end and allows this strand to be elongated. Ultimately, DNA Pol a/primase can synthesize a primer on this strand, which is elongated by DNA Pol . Some organisms rely on a different mechanism for replication of telomeric DNA. For example, flies lack telomerase and maintain telomere length by regulated insertion of non-LTR retrotransposons into telomere DNA.