ppt presentation
advertisement

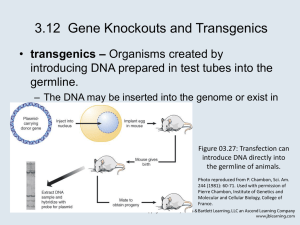
Transgenosis methods of transformation Foreign DNA introduction • transient DNA is / is not integrated to chromosom/plastom/chondriom) • stable Stable integration results in: • mutation (always, but negative effects are rare) • expression of introduced gene (if intended, allways unsure – RNAi, …) Expression of introduced gene - universality of genetic code allows functional protein synthesis even from genes of distant organisms - specific regulatory sequences necessary for transcription and translation PROMOTER: - start site and direction of transcription - interacts with trans elements (transcription factors = specific spatiotemporal regulation of activity – specific host promoters) - special promoters: constitutive, inducible (EtOH, heat, estradiol) - origin: plants, plant pathogenes, synthetic TERMINATOR - balanced with promoter! Translation AACAATG Stop mRNA coding sequence - codon usage can differ (tRNA frequency) - problematic if expressing plant gene in bacteria - ATG sequence context – important!!! („Kozak sequence“) - posible problems with splicing (differences animal – plant) Preparation of transgenic plant – general procedure 1. One transformed cell 2. Multiplication (usually under selection) – callus formation 3. Induction of organogenesis (somatic embryogenesis) - plant growth regulators DNA insertion selection organogenesis embryogenesis transformed cell transformed callus Selection of transformed cells (plants) Selection genes resistances (degradation/modification of selection agens or production of insensitive target) - antibiotics (kanamycin, hygromycin) - herbicides (Roundup® - glyphosate, Liberty®=basta – glufosinate=phosphinotricin) other, e.g. PMI (phosphomanose isomerase) - conversion of manose-6-P na fru-6-P Reporter genes - visual selection (GFP, GUS, …) Plant cell transformation methods „natural“ method • via Agrobacterium – modifications (agroinfection, vacuum infiltration) • with plant virus – transient transformation (DNA not integrated) – often primary infection with DNA copy of the genome (via Agrobacterium) biolistics („particle bombardment“, „microprojectile bombardment“) „direct gene transfer“ to protoplast • electroporation • polyethylenglycol (PEG) microinjection Natural transformation with agrobacterium (Agrobacterium tumefaciens) • soil bacteria , G- (Rhizobiaceae), Ti plasmid • „genetic parsitism“ in dicots (external activation of monocots with acetosyringon) • transfers several genes within T-DNA to plant cell (causing tumor formation from transformed cell) ipt – isopentenyl transferase iaaH – indolacetamid hydrolase genes for opine synthesis Natural transformation with agrobacterium Disarmed agrobacterium - tumor and opine genes removed – only border sequences necessary for T-DNA mobilization - Ti plasmid splited into two plasmids: • T-DNA in small binary vector • vir genes in helper plasmid Border sequences: - imperfect direct repeat 25 bp: (LB a RB = right, left border) - single strand breakage (virD2) (D1/D2 dimer), strand replacement Vir proteins in T-DNA transfer induction of vir region expression in reaction on phenolic compound (VirA,VirG) cleavage at T-DNA ends (VirD1, VirD2) porus formation (VirB1-11) protection of ssT-DNA (VirE2) transfer of T-DNA to nucleus (VirE2 binds transcription factor VIP, VirD2 contains NLS – interaction with importin KAP-α) targeted proteolysis of T-DNA complex before integration (VirF) Integration of T-DNA - non-homologous recombination - microhomology of inserted sequence in integration site - often short deletions, rearangements, filler sequence - RB more frequently preserved VirD2 protein) Alternatively: formation of dsT-DNA, ligation into ds break of chromosome Advantages of agro-transformation • relatively high frequency of stable transformation • low copy number (lower risk of RNAi induction) • relatively long sequences (up to 45 kbp) Transformation procedures: • simple cocultivation of agro with plant tissue, cell culture, …. • vacuum infiltration of agro to the tissue • inoculation in planta (flowers, leaves, …) Potato transformation with agrobacterium agrobacterium entrance to the tissue microscopical callus agrobacterium 3-4 weeks cocultivation with injured leaves 5-6 weeks Floral dip in Arabidopsis thaliana inoculation in planta Inflorescences with buds into agrobacterium suspension Floral dip in Arabidopsis thaliana inoculation in planta - target structure mostly the egg – forms transformed embryo - plant GUS reporter in developing transformed seeds Agroinfiltration of tobacco (in planta) - preliminary tests of transgene expression - without in vitro work - cotransformed with viral supressor of silencing (p19) to reduce PTGS + P19 without P19 Biolistic method (Partickle gun, Gene gun) • coating of Au or W particles with DNA • shot onto the tissue Biolistics over- or under-pressure to mobilize particles Biolistics • transformed cell through regeneration • universal usage without limitations (only regeneration ability) • transformation of organelles (chloroplasts) gold particle (1 m) chloroplast (5 x 3 m) Chloroplast transformation - higher copy number per cell - high protein levels - without silencing - targeting within plastom possible thanks to active homologous recombination - plastid genome usually missing in pollen Disadvatages: - no eukaryotic posttranslational modifications - preparation of homoplastic and non-chimeric plants is time consuming Chloroplast transformation Integration by recombination – transgene surrounded with plastid sequences Gradual selection of homoplasmic non-chimeric plants Viral vectors for transient biotechnological expression of proteins - episomal - non-integrated (= no position effect) - high copy number - strong expression – fast accumulation of product - natural supressors of silencing (x PTGS) - systemic spreading within plant - often wide host spectrum Viral vectors - substitutional (e.g. for capsid protein, in viruses with polyhedral capsids) - insertional (possible in helical viruses) - modular – splitting into more replicons (e.g. TMV, Geminiviridae – polyhedral capsid) Viral vectors originally from Caulimoviridae - dsDNA genome – enabled sequence modification low capacity (to 500 bp – polyhedral capsid) polycistronic transcripts (complicated changes) no practical application at present: - - helical viruses (TMV) tolerant to longer inserts RNA viruses (enabled by discovery of RT – modification of cDNA of viral genome) primary infection in DNA form (agrobacterium) – viral genome by transcription Transgene incorporation - ssT-DNA: non-homologous recombination - dsDNA: reparation of DSB (double stranded break of DNA) • simple joining of DNA ends – blunt ends (frequent deletions) – dominant in plants (most Eucaryots) • homologous recombination – integration in yeast, plastids and Physcomitrella – „repair“ according to template (sister chromatid, inserted sequence) Homologous recombination – basic function • crossing-over in meiosis (homologous chromosoms) • DNA repair (sister chromatid) Homologous recombination 1. extrachromosomal recombination - between two introduced molecules - frequency in plants: 1 - 4 % 2. Intrachromosomal recombination - between two loci in the same chromosome - frequency in plants: 10-5 až 10-6 3. gene targeting recombination between introduced and chromosomal DNA CHS lokus (targeted integration of transgene in yeast, plastids, Physcomitrella, …) NPTII in other eucaryots can be induced by formation of DSB in the target sequence! vektor pro gene targeting oblasti homologie upravený CHS lokus NPTII Frequency of homologous recombination (after insertion of homologous sequence) ratio between homologous/non-homologous recombination Higher plants 10-3 - 10-6 (high frequency of non-homologous recombination prevets homologous) mammels 10-2 -10-5 lower eucaryots (yeast, protists, filamentous fungi) > 10% moss Physcomitrella patens cca 90% - effected by sequence length, ploidy, cell type, cell cycle phase, … Stimulation of targeted DNA insertion homologous recombination: - DSB formation through site specific nuclease (ZFN, TALEN, CRISPR/Cas) – integration by both homologous and non-homologous recombination non-homologous recombination - site specific rekombination systems of prokaryots and low eucaryots (recombinase/recognized target site – has to be introduced to the genome in advance) Cre/lox bacteriophage P1 Flp/frt Saccharomyces cerevisiae R/RS Zygosaccharomyces rouxii Cre/loxP - Integration to previous insertion site - Specific removal of selection marker gene Targeted formation of DSB Cleavage of unique genomic sequence that is recognized - by chimeric nucleases, whose DNA binding domain can be suited for specific sequence: Zn-finger nucleases (ZFN) TALEN (transcription activator-like effektor nucleases - by complementary RNA CRISPR/Cas Repair of DSB Connected with DNA insertion Without DNA insertion - integrace transgenu s homologními koncovými sekvencemi (homologní rekombinací) - autonomous repair often results in local deletion - integrace nehomologní rekombinací v místě DSB - presence (insertion) of DNA template can direct specific site mutagenesis by homologous recombination Zn-finger domains – designed for specific sequence DNA sequence recognition: 3(-4) nt/finger - conserved aminoacids marked - black circles – interaction with DNA - místně specifické štěpení - využití k cílené modifikaci DNA (integraci transgenu)