DNA Recombination

DNA Recombination
• Roles
• Types
• Homologous recombination in E.coli
• Transposable elements
Biological Roles for Recombination
1. Generating new gene/allele combinations
(crossing over during meiosis)
2. Generating new genes (e.g., Immunoglobulin rearrangement)
3. Integration of a specific DNA element (or virus)
4. DNA repair
Practical Uses of Recombination
1. Used to map genes on chromosomes
- recombination frequency proportional to distance between genes
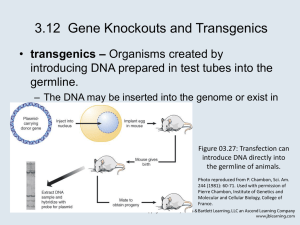
2. Making transgenic cells and organisms
Map of Chromosome I of
Chlamydomonas reinhardtii
cM = centiMorgan; unit of recombination frequency
1 cM = 1% recombination frequency
Chlamydomonas Genetics Center
Types of Recombination
1. Homologous - occurs between sequences that are nearly identical (e.g., during meiosis)
2. Site-Specific - occurs between sequences with a limited stretch of similarity; involves specific sites
3. Transposition – DNA element moves from one site to another, usually little sequence similarity involved
Examples of (mostly) Homologous Recombination Fig. 22.1
Holliday Model
R. Holliday (1964)
Holliday Junctions form during recombination
- HJs can be resolved
2 ways, only one produces true recombinant molecules patch
EM of a Holliday Junction w/a few melted base pairs around junction
Fig. 22.3
The recBCD
Pathway of
Homologous
Recombination
Part I : Nicking and
Exchanging
Fig. 22.2 a-d
recBCD Pathway of Homologous Recomb.
Part I: Nicking and Exchanging
1.
A nick is created in one strand by recBCD at a Chi sequence (GCTGGTGG), found every 5000 bp.
2.
Unwinding of DNA containing Chi sequence by recBCD allows binding of SSB and recA.
3.
recA promotes strand invasion into homologous
DNA, displacing one strand.
4.
The displaced strand base-pairs with the single strand left behind on the other chromosome.
5.
The displaced and now paired strand is nicked
(by recBCD?
) to complete strand exchange.
recBCD Pathway of Homologous
Recombination
Part II : Branch
Migration and
Resolution
Fig. 22.5 f-h
recBCD Pathway of Homologous Recom.
Part II: Branch Migration and Resolution
1. Nicks are sealed Holliday Junction
2. Branch migration ( ruvA + ruvB )
3. Resolution of Holliday Junction ( ruvC )
RecBCD : A Complex Enzyme
• RecBCD has:
1. Endonuclease subunits ( recBC ) that cut one DNA strand close to Chi sequence.
2. DNA helicase activity ( recD subunit) and a DNA-dependent ATPase activity
– unwinds DNA to generate the 3’ SS tails
RecA
• 38 kDa protein that polymerizes onto SS DNA 5’-3’
• Catalyzes strand exchange, also an ATPase
• Also binds DS DNA, but not as strongly as SS
RecA binds preferentially to SS DNA and will catalyze invasion of a DS
DNA molecule by a SS homologue.
Important for many types of homologous recombination, such as during meoisis (in yeast).
Fig. 6.19 in Buchanan et al.
RecA Function Dissected
• 3 steps of strand exchange:
1. Pre-synapsis : recA coats single-stranded
DNA (accelerated by SSB, so get more relaxed structure).
2. Synapsis : alignment of complementary sequences in SS and DS DNA (paranemic or side-by-side structure).
3. Post-synapsis or strand-exchange : SS DNA replaces the same strand in the duplex to form a new DS DNA (requires ATP hydrolysis).
RuvA
and
RuvB
• DNA helicase that catalyzes branch migration
• RuvA tetramer binds to HJ (each DNA helix between subunits), forces it into square planar conformation
• 2 copies of RuvB bind at the HJ (to RuvA and 2 of the DNA helices)
– RuvB is a hexamer ring, has helicase & ATPase activity
• Branch migration is in the direction of recA mediated strand-exchange
RuvB
RuvA/RuvB/DNA Complex
RuvA
Shows RuvB encircling DNA duplexes
RuvA
RuvB
RuvA removed for visual purposes only
Similar to Figure 22.13 Model based on EM images.
RuvC
: resolvase
• Endonuclease that cuts 2 strands of HJ
• Binds to HJ as a dimer (that already has
RuvA/RuvB)
• Consensus sequence: (A/T)TT (G/C)
- occurs frequently in E. coli genome
- branch migration needed to reach consensus sequence!
RuvC bound to a HJ
Fig. 22.16
A model for binding of RuvA, RuvB, and RuvC to a HJ.
Fig. 22.17b
Meiotic
Recomb. in
Yeast
- is initiated by a double-strand break (DSB)
Fig. 22.18
Repair of double-strand breaks (DSBs) in non-dividing or mitotic cells
DSBs probably most severe form of DNA damage, can cause loss of genes or even cell death (apoptosis)
DSBs caused by:
- ionizing radiation
- certain chemicals
- some enzymes (topoisomerases, endonucleases)
- torsional stress
2 general ways to repair DSBs:
1.
Homologous recombination (HR) - repair of broken
DNA using the intact homologue, very similar to meiotic recombination . Very accurate.
2.
Non-homologous end joining (NHEJ) - ligating nonhomologous ends. Prone to errors, ends can be damaged before religation (genetic material lost) or get translocations. ( Mechanism in Fig 20.38
)
Usage: NHEJ >> HR in plants and animals