Diagnostic and prognostic tools in haematological malignancies in
advertisement

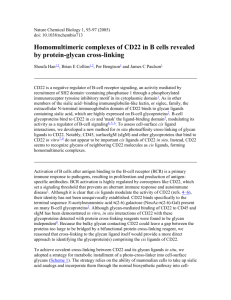
DIAGNOSTIC AND PROGNOSTIC TOOLS IN HAEMATOLOGICAL MALIGNANCIES IN 2010 (PART II) Flow cytometry Immunophenotyping • Identification of molecular protein characteristics of abnormal cells • Labelling with fluorescent monospecific monoclonal antibodies • Multiparametric flow cytometry The objectives of immunophenotyping in hematological disorders • Lineage assignment – Lymphoid acute/chronic – Myeloid acute/chronic – Erythroid – Megakaryocytic • Classification /Scoring • Detection of MPAL • Maturation anomalies • Identification of Minimal Residual Disease patterns Before.... Now..... Harrogate 7th HLDA : 250 CD Adelaide 2004 8th HLDA : 339 CD Barcelona 2010 : >360 CD Immunophenotype of lymphoid cells • Well established maturation sequence • Staged and controlled to prepare long lived antigen-specific cells • Identified by studying leukemias • Validated on normal cells • Numerous similarities between B and T cells: – Lineage committment – Gene rearrangements for antigen recetpors – Selection B lineage associated markers CD22 or CD19 BCR CD79 CD21 ITAM ITAM ITAM ITAM ITIM ITAM ITIM ITAM ITIM ITIM CD23 COOH CD20 CD24 P NH2 BONE MARROW Stem Cell Pro-B Cell Early-B Cell Pre-B Cell Naive B cell H0L0 H0L0 TISSUES ACTIVATION & CLONAL PROLIFERATION Immunocyte Immunoblast HxL0 HxL0 HRL0 HRLR CD34 cCD79 cCD79 cCD79 cCD79 sCD79 DR cCD22 CD22 CD22 CD22 CD22 CD19 CD19 CD19 CD19 CD19 CD21 CD20 CD20 CD20 CD10 CD21 CD21 CD21 CD10 cµ sµ/sd Memory B cells Plasma cells Classification of B- ALL (EGIL) cCD79/CD 19/c or sCD22 CD10 c-µ sIg B-I (pro-B) + - - - B-II (common) B-III (pre-B) + + - - + + + - B-IV (mature) + + + + T lineage associated markers TCR CD3g d e ITAMITAMITAM CD5 ezz/zh CD2 ITAMITAMITAM ITAMITAM ITAMITAM CD4 CD1 CD8 CD7 BONE MARROW Corticothymocyte Stem cell Pro-T cell TISSUES THYMUS Medullary Thymocyte sCD3 CD7 CD2 CD5 CD8 Naïve T cell ACTIVATION & CLONAL PROLIFERATION Immunocyte Immunoblast g0d0 a0b0 CD34 DR g0d0 a0b0 cCD3 CD7 CD2 CD5 gXdX a0b0 cCD3 CD7 CD2 CD5 CD1 gXdX aXbX cCD3 CD7 CD2 CD5 CD4 CD8 gXdX aRbR sCD3 CD7 CD2 CD5 CD4 Memory T cells Effector T cells Classification of T-ALL (EGIL) cCD3 CD7 CD2 CD5 CD8 CD1a+ sCD3+/ CD1a- T-I (pro-T) + + - - - T-II (pre-T) + + + - - T-III (cortical T) T-IV (mature T) + + + + - + + + - + Lymphoproliferative disorders MARGINAL ZONE NODAL MALT T LYMPHOMAS SEZARY) LARGE CELLS ANAPLASTIC PERIPHERAL ANGIOCENTRIC INTESTINAL HCL MANTLE ZONE BURKITT INTESTINAL OR LUNG MALT Follicular, mantle zone, marginal zone SLVL FOLLICULAR WALDENSTROM CENTROBLASTIC or IMMUNOBLASTIC Large cell Lymphomas Circulating marginal zone cells MYELOMA Matutes’ scoring of CLL POSITIVE NEGATIVE OR WEAK CD5 1 0 CD23 1 0 sIg 0 1 FMC7 0 1 CD22/CD79b 0 1 Comparative immunophenotypes SMZL/SLVL HCL HCLv CD19 CD22 CD5 CD23 sIg CD43 FMC7 CD10 CD103 CD11c CD25 CD79b CD20 IgM IgM, IgG.. IgG CLL MCL FL Immunophenotype of myeloid cells • Known maturation sequence • Massive production of cells with no specificity • Early and late differentiation markers • Important lineage promiscuity Myeloid lineage markers CD117 MPO CD13 CD33 COOH E E CD35 NH2 CD14 P L L L L L L L L L L K CD11b Mg Mg Mg CD15 CD65 CD36 3-fucosyl-N-acetyl-lactosam céramide dodecasaccharid DR BONE MARRROW PERIPHERAL BLOOD TISSUES CD4 CD19 CD11B CD14 CD36 CD34 DR Stem cell CD117 CD13 CD33 MPO CD7 Immature precursors CD15 CD65 CD16 CD32 CD64 Differentiated cells Evolution immunohénotypique au cours de la maturation granuleuse (B Husson) Myeloblast CFU-GM Promyelocyte Myelocyte Metamyelocyte Band Granulocyte CD15 CD66 HLA-DR CD11b CD16 CD117 CD34 CD33 CD13 CD13 CD11c CD11c CD33 CD15 CD11b CD66 CD16 Detection of BAL : EGIL’scoring system • NOT to be used for lineage assignment • Scoring based on the lineage specificity of critical differentiation antigens • Calculation of each lineage “score” • In BAL, at least two lineages have scores HIGHER than 2 • In “variant” AL coexpression with a score <2 is not rare Detection of BAL : EGIL ’scoring system BLINEAGE T-LINEAGE MYELOID LINEAGE 2 points CD79 cµ cCD22 CD3 TCR MPO (lysozyme) 1 point CD19 CD10 CD20 CD2 CD5 CD8 CD10 CD13 CD33 CDw65 CD117 0.5 point TdT CD24 TdT CD7 CD1a CD14 CD15 CD64 WHO’s new classification Extensive immunophenotype AUL Acute undifferentiated leukemia With t(9;22) Others MPAL Incl NK Mixed Phenotype Acute leukemia With t(n;11q23) NOS WHO’s new criteria of MPAL • Myeloid lineage – MPO – Or strong monocytic engagement • NSE • CD14, CD11c, CD36, CD64, Lysozyme • B-lineage – Bright CD19 + another B marker – Low CD19 + two other B markers • T-lineage : cCD3 (strong, PE or APC) One word on MDS Proposition de score Wells, 2003 0 1 2 3 Working conference 2008 M Loken, A van de Loosdrecht, K Ogata, A Orfao, D Wells • Classical blasts enumeration – DR+/11b– CD34 – CD117 • Anomalies of the CD13/CD16 pathway • Abnormal expression anormale of DR on monocytes Stachurski, 2008 • Blasts – – – • Granulocytes – – • CD34, CD117 Abnormal CD2, CD5, CD7, CD56 Increased CD117 Deganulation Abnormal expression of CD33, CD13, CD11b, CD16, CD15, CD64, CD10, CD14, DR Monocytes – Anomalies of CD33, CD13, CD11b, CD15, CD64, CD14, DR In practice, for hematological malignancies • Depending on – Previous clinical and morphological information – Sample volume – Monoclonal antibodies availability Consensual European Panel, European LeukemiaNet (2005) For quick orientation or paucicellular samples • cCD3, MPO, cCD79a, TdT • CD7, CD2, CD10, CD19, CD22 (s or c), sIg, CD13, CD33, CD34 • CD45 for gating purposes Sublineage classification and definition of clinical entities (also with adapted gating strategy) • DR, CD1a, CD4, CD5, CD8, CD3 (m), IgM (c), CD14, CD117, CD56, CD65, CD41 or CD61, RBC marker such as glycophorin A Complementary panel of useful referenced markers Acute leukemia Diagnosis Panel Other useful markers (>20) • • • • • • • • • • • • • • • • MPO/LF (lactoferrin) (c): (i) Identification of late neutrophil granulocyte compartment (Lactoferrin positive) (25,52,54); (ii) for refined detection/quantification of MPO+ early myeloid cells in combination with CD14 (25,49) LZ (lysozyme) (c): (i) for myeloblastic leukaemia; (ii) to discriminate pDCs and myeloid cells (53); (iii) to positively identify early monocytic cells (48) K/L : (i) on surface for clonality, (ii) in the cytoplasm for rare B-IV cases CD11b, CD11c : negative in APL (14) CD15 : for myeloblastic leukemia (42) CD16 : to discriminate mature PMNs (9) CD35/36 : for GEIL´s AML classification (11), for RBC after excluding monocytes (10) CD58 : to distinguish between normal regenerating B cells and B-cell blasts (59) CD64 : for AML (9) CD68 (c): (i) for AML (bright) and subset of B-ALL (weak) (53); (ii) for positive identification of normal pDCs (bright) (55) CD71 : for cell proliferation/activation and/or RBC (22,41) CD86 : prognostic factor in AML (34) CD99 : to differentiate between blasts and non blastic T-cells (19) CD123 : IL-3 R, for pDC and AML, some NK (24) TCR chains for T-ALL, c and/or s (50) Therapeutic targets: CD20 (40), CD52 (40), CD45 (39), CD33 (31), CD123 (3), CD87 (46), CD44 (20), uPAR(CD87)/uPACD116 (1) Chronic lymphoproliferative diseases. Mandatory panel (20 Abs) • Samples : peripheral blood, bone marrow, LN suspensions… (Fine needle aspiration for primary screeening to avoid unnecessary biopsies) • Gating markers CD19, CD3, CD56* • B oriented panel gated on CD19 CD5, CD20, CD23, CD103, CD10, K, L, Ig, CD25, CD79b, CD38 • T oriented panel gated on CD3 or other T-lineage marker CD2, CD3, CD4, CD5, CD8, CD7, Lymphoproliferative diseases: additional useful markers (< 15) • B lineage – CD2, CD7, CD123, FMC7, CD138, DR, CD24*, CD43 – (G, A, M, D), CD81* – Cytoplasmic : Bcl2, Zap70 (relative to internal control) • T lineage – TCRs*, CD30, CD10 • NK panel excluding CD19 and CD3 cells/ CD56 – CD57, CD16, CD94, perforin, granzyme B *CD81/CD22 useful for CLLfollow-up based on dim co-expression level *V-beta panel and imunophenotype *Absence of CD24 on marginal zone and HCL • Note : CD52 if alemtuzumab considered Chemosensitivity • Minimal residual disease • Quick assessment of response to therapy Why look for minimal residual disease? The founders San Miguel, 1997 Campana, 1999 Kern, 2004 MRD in ALL MRD B-II ALL CD34 1.5 x 10-2 CD19 MRD in AML CD117 MRD - AML CD34 2.1 10-3 AML peripheral blast cells decrease D0 SSC SSC A a CD45 CD16 CD16 e D CD45 5.3% Blasts 100 g CD11b h CD11b CD16 CD11b SSC d CD16 SSC CD14 b C B c FSC f E CD45 3.9% Blasts 73.7 b CD11b e D2 CD16 CD16 D1 CD11b 1.1% Blasts CD11b CD16 g 0.5% 0.2% Blasts 9.4 f CD11b D4 D3 21.2 d CD11b CD16 c CD16 CD11b CD16 CD16 CD16 a CD11b Blasts 3.6 h CD11b Blast slope 100 90 80 70 DFS 60 50 Slope < -25 40 Slope 30 -25 < > -15 Slope > -15 20 10 0 0 200 400 600 800 Time (Days) 1000 1200 1400 Conclusions • • • • • Consensual approach Rapid and informative diagnostic tool Precise definition of the disease at diagnosis Therapeutic indications Aberrant immunophenotypes useful for follow up • Search for new markers: – New monoclonals – Microarrays