Transcription and Translation - Microbiology and Molecular Genetics
advertisement
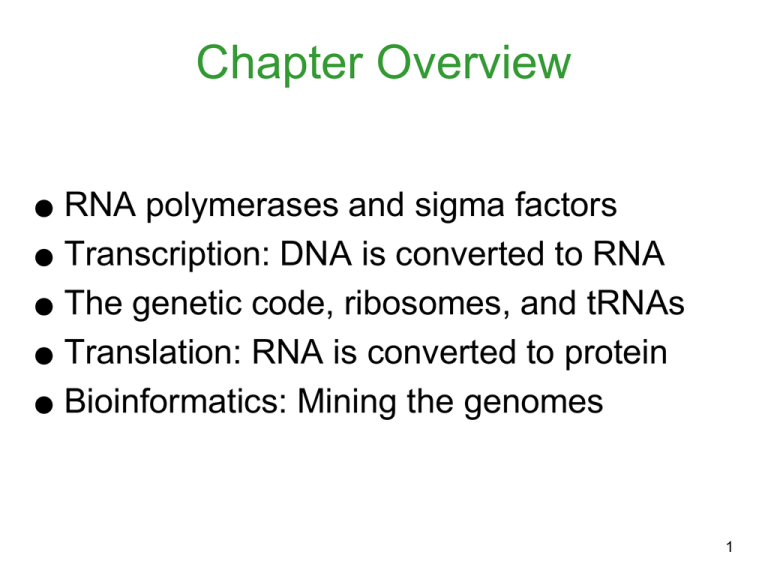
Chapter Overview RNA polymerases and sigma factors ● Transcription: DNA is converted to RNA ● The genetic code, ribosomes, and tRNAs ● Translation: RNA is converted to protein ● Bioinformatics: Mining the genomes ● 1 Introduction The cell accesses its vast store of data in its genome by: - Reading a DNA template to make an RNA copy (transcription) - And decoding the RNA to assemble protein (translation) After translation, each polypeptide is properly folded and placed at the correct cellular or extracellular location. 2 RNA Polymerase Is a complex enzyme that carries out transcription by making RNA copies (called transcripts) of a DNA template strand In bacteria, the RNA pol holoenzyme is composed of: - Core polymerase: a2, b, b´ - Required for the elongation phase -Holoenzyme: s , a2, b, b´ - Sigma factor: s - Required for the initiation phase 3 RNA Polymerase • RNA poymerase links nucleotides in the 5’ 3’ • Opens DNA by itself (helicase is not required) • Transcription is slower than replication (~ 50 nucleotides/sec) • Lacks proofreading function (errors 10-4). 4 Figure 8.2 Figure 8.3 5 The sigma factor helps the core enzyme detect the promoter, which signals the beginning of the gene. Every cell has a “housekeeping” sigma factor. - In E. coli, it is sigma-70. - Recognizes consensus sequences at the –10 and –35 positions, relative to the start of the RNA transcript (+1) A single bacterial species can make several different sigma factors. 6 How Sigma Factor Recognizes Specific DNA Sequences Orientation of the promoter determines the direction of the transcription 7 Alignment of sigma -70 (s) dependent promoters from various genes is used to generate consensus sequences. Yellow= conserved region; Brown= transcript start site. 8 Transcription of DNA to RNA Transcription occurs in three phases: 1) Initiation: RNA pol holoenzyme binds to the promoter - The closed RNA pol complex becomes open. 2) Elongation: The RNA chain is extended 3) Termination: RNA pol detaches from the DNA, after the transcript is made 9 10 Transcription Initiation • Transcription can occur either strands • Only one DNA strand is transcribed (sense strand) • Transcription proceeds 5’ 3’ • The first base is usually a purine (A or G) added to the +1 site. • Orientation of the promoter determines the direction of the transcription 11 Energy released in this process is used to build phosphodiesterase bonds 12 Transcription Elongation • Is the sequential addition of ribonucleotides • • • from nucleoside triphosphates The original RNA polymerase continues to move along the template, synthesizing RNA at ~ 45 bases/sec. The unwinding of DNA ahead of the moving complex forms a 17-bp transcription bubble. Positive supercoils ahead are removed by DNA topoisomerases. 13 Now we have some idea of how RNA polymerase recognizes the beginning of a gene and how the transcription proceeds! But how does it know when to stop 14 The secret is in the sequence ! 15 Transcription Termination There are two types of transcription: - Rho-dependent - Relies on a protein called Rho and a strong pause site at the 3´ end of the gene - Rho-independent - Requires a GC-rich region of RNA, as well as 4–8 consecutive U residues 16 Figure 8.8 17 Termination of transcription DNA 5’ Promoter Operator A Transcription B Terminator C 3’ 18 Antibiotics that Affect Transcription Rifamycin B - Selectively binds to the bacterial RNA pol - Inhibits transcription initiation Actinomycin D - Nonselectively binds to DNA - Inhibits transcription elongation 19 Six Classes of RNA Messenger RNA (mRNA): Encodes proteins Ribosomal RNA (rRNA): Forms ribosomes Transfer RNA (tRNA): Shuttles amino acids Small RNA (sRNA): Regulates transcription or translation tmRNA: Frees ribosomes stuck on damaged mRNA Catalytic RNA: Carries out enzymatic reactions 20 Translation: mRNA Protein mRNA contains codes for how to make a proteins ! 21 The Genetic Code Consists of nucleotide triplets called codons There are 64 possible codons: - 61 specify amino acids. - Include the start codons (AUG) - 3 are stop codons (UAA, UAG, UGA) The code is degenerate or redundant. - Multiple codons can encode same amino acid. The code operates universally across species. - Remarkably, with very few exceptions 22 Figure 8.11 23 The Genetic Code • Degeneracy: redundancy (e.g. leucine has 6 codons and alanine has 4 codon) 24 tRNA Molecules Are decoder molecules that convert the language of RNA into that of proteins tRNAs are shaped like a clover leaf (in 2-D) and a boomerang (in 3-D). A tRNA molecule has two functional regions: - Anticodon: Hydrogen bonds with the mRNA codon specifying an amino acid - 3´ (acceptor) end: binds the amino acid 25 Figure 8.12B -About 60 different t-RNAs in bacteria -About 20 aminoacyl-tRNA synthetases Figure 8.13 26 Figure 8.15 The charging of tRNAs is carried out by a set of enzymes called aminoacyltRNA synthetases. 27 The Ribosome • Ribosomes are composed of two subunits, each of which includes rRNA and proteins. • In prokaryotes, the subunits are 30S and 50S and combine to form the 70S ribosome. • The 30S contains 21 proteins (S1-S21) assembled around 16S rRNA • The 50S contains 31 proteins (L1-L31) associated with 5S and 23 S rRNA 28 29 The 70S ribosome harbors three binding sites for tRNA: - A (acceptor) site: Binds incoming aminoacyl-tRNA - P (peptidyl-tRNA) site: Harbors the tRNA with the growing polypeptide chain - E (exit) site: Binds a tRNA recently stripped of its polypeptide 30 Translation of RNA to Protein Polypeptide synthesis occurs in 3 phases: 1) Initiation: which brings the two ribosomal subunits together, placing the first amino acid in position 2) Elongation: which sequentially adds amino acids as directed by mRNA transcript 3) Termination: which releases the completed protein and recycles ribosomal subunits Each phase requires a number of protein factors and energy in the form of GTP. 31 How do ribosomes find the right Reading Frame? 32 Defining a Gene Alignment of a bacterial structural gene with its mRNA transcript Figure 8.21 33 Open Reading Frames (ORF) mRNA sequence AUG GCA UUG CCU UAG Start -------------------------Stop Reading Frame # 1 AUG GCA UUG CCU met ala leu pro Reading Frame # 2 A UGG CAU UGC CU try his cys Reading Frame # 3 AU GGC AUU GCC U gly Ile ala 34 Translation Initiation Figure 8.23 35 Translation Elongation` Three steps are repeated: • t-RNA-carrying an amino acid binds to “A” site • peptide bond formation occurs • the message must move by one codon 36 Translation Termination 37 38 Coupled transcription and translation in prokaryotes. 39 Antibiotics that Affect Translation Streptomycin: Inhibits 70S ribosome formation Tetracycline: Inhibits aminoacyl-tRNA binding to the A site Chloramphenicol: Inhibits peptidyltransferase Puromycin: Triggers peptidyltransferase prematurely Erythromycin: Causes abortive translocation Fusidic acid: Prevents translocation 40 Protein Modification Protein structure may be modified after translation: - N-formyl group may be removed by methionine deformylase. - The entire methionine may be removed by methionyl aminopeptidase. - Acetyl groups or AMP can be attached. - Proteolytic cleavages may activate or inactivate a protein. 41 What is bioinformatics? Bioinformatics is the field of science in which biology, computer science, and information technology merge to form a single discipline. The ultimate goal of the field is to enable the discovery of new biological insights as well as to create a global perspective from which unifying principles in biology can be discerned 42 Bioinformatics Since 1998, the complete genomes of more than 225 microbial species have been published. This wealth of information has spawned a new discipline called bioinformatics, which is dedicated to comparing genes of different species. Data from bioinformatics enable scientists to make predictions about an organism’s physiology and evolutionary development. - Even without culturing the organism in a lab 43 Annotating the Genome Sequence Annotation of the DNA sequence is basically understanding what the sequence means. - It requires computers that look for patterns, such as regulatory sequences, open-reading frames (ORFs), and rDNA and tRNA genes An ORF is a sequence of DNA that encodes an actual polypeptide. - In eukaryotes, finding ORFs is complicated by the presence of introns. 44 DNA Sequence >A01_TK-M13F-Plate5.ab1 1360 0 1360 ABI TTCCTAAGCTGGTTACTAGACTGCACATTGGGCCCTCTAGAGATGCTCGAGCGGCCGCCAGTGTGATGGATATCTGCAGAATT CGCCCTTGTGCCAGCCGCCGCGGTAATACGTAGGGCGCAAGCGTTGTCCGGAATTATTGGGCGTAAAGAGCTCGTAGGCGG CTTGTCGCGTCGGTTGTGAAAGCCCGGGGCTTAACCCCGGGTCTGCAGTCGATACGGGCAGGCTAGAGTTCGGTAGGGGAG ATCGGAATTCCTGGTGTAGCGGTGAAATGCGCAGATATCAGGAGGAGCACCGGTGGCGAAGGCGGATCTCTGGGCCGATACT GACGCTGAGGAGCGAAAGCGTGGGGAGCGAACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGGTGGGCACTAGGTG TGGGCCACATTCCACGTGGTCCGTGCCGCAGCTAACGCATTAAGTGCCCCGCCTGGGGAGTACGGCCGCAAGGCTAAAACTC AAAGGAATTGACGGGGGCCCGCACAAGCGGCGGAGCATGTGGCTTAATTCGACGCAACGCGAAGAACCTTACCAAGGCTTGA CATACACCGGAAACATTCAGAGATGGGTGCCCCCTTGTGGTCGGTGTACAGGTGGTGCATGGCTGTCGTCAGCTCGTGTCGT GAGATGTTGGGTTAAGTCCCACAACGAGCGCAACCCTTGTCCCGTGTTGCCAGCAGGCCCTTGTGGTGCTGGGGACTCACGG GAGACCGCCGGGGTCAACTCGGAGGAAGGTGGGGACGACGTCAAGTCATCATGCCCCTTATGTCTTGGGCTGCACACGTGCT ACAATGGCCGGTACCATGAGCTTCCATACCGCAAGGTGGAGCGAAACTCAAAAAGCCGGTCTCACTTCCGATTGGGGTCTCC ACCTCCCCCCCCTGCAATTTGATCCCGTGTAATACTGGATATAAGTGTTGCGGGGAAACCTTCCCGGGGGTGTTTACCCCCCC CTTCAAGAGGGAATTCCTCCCAACCGGCGGCGCCTTTCTAGTGAGAACCCACCCGTGTGCCAACCTTTGATTAATTTATGGGG GGTTGTTTTTTTTATTAACAAAGNNNNNGTNACANNGGNNAANCGCCCCGGGGCCGTTCACCCCCCCTATAATTGCCCTTTGTT GACGAATTACCCCCCTTTTCGCCCGTGGTCCGCGACCCCAAATACCCCACAAGCAGGTCCCAGCCCACCCAATTCCCCCATG TCCCCCCCCATCCCCCTCGTCTTCTTAACCTTCGCGCCGAGTGGTGTTAAACAGGGGAGGTCCGCGCTGGATATCGTTTTTTT TGATGTTATGGCAGCTCCTCCTAGATTTATAGACGCCCCCCGCG 45 Predicting Open Reading Frames (ORFs) in a DNA sequence Predicting a Open Reading Frame (ORF). Prediction begins locating the: -Start codon: AUG in m-RNA (TAC on sense DNA). -Stop codons: UAA, UAG, and UGA in m-RNA (ATT, ATC, and ACT on sense DNA) -Ribosome-binding site: upstream of start the start codon 46 Mycoplasma mycoides: Color code indicates gene clustering by function. The inner most circle shows GC content. Red , > 50% and black, < 50%. 47 Evolutionary Relationships Genes that are homologous likely evolved from a common ancestral gene. - Orthologous genes - Genes duplicated via appearance of a new species - Have identical function in different organisms - Paralogous genes - Genes duplicated within a species - Have slightly different tasks in a cell 48 49 Bioinformatics Many computer programs and resources used to analyze DNA and protein sequences are freely available on the Web. - BLAST - Multiple Sequence Alignment - KEGG - Motif Search - ExPASy - Joint Genome Institute 50