EPIGENETIKA
SUMMER 2011
Petr Svoboda
mail:
tel:
svobodap@img.cas.cz
241063147
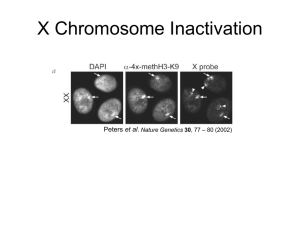
X-INACTIVATION
Costs and benefits of sex
“The outstanding puzzle of evolutionary biology”
Sex is very costly - A population of sexually-reproducing organisms
has 50% of fitness (reproductive rate) compared to asexually
reproducing population of the same size. Still, the majority of
multicellular organisms reproduces sexually.
Sexual reproduction must have some compensating advantage
- sex can accelerate the rate of evolution (combination of good traits)
- sex offers increased variability (gene pools)
- genome maintenance (reduction of deletetious mutations)
A population of sexually reproducing organisms can, under some
conditions, evolve faster than a similar number of asexual organisms.
• If favorable mutations arise more frequently,
Fisher's argument works: the sexual population
evolves faster. Each new favorable mutation will
usually arise in an individual that does not already
possess other favorable mutations; the greater
speed with which the different favorable mutations
combine together causes the sexual population to
evolve faster.
• If favorable mutations are rare, each one will have
been fixed in the population before the next one
arises. New favorable mutations will always arise in
individuals that already carry the previous favorable
mutation. Sexual and asexual populations then
evolve at the same rate.
a.k.a. Fisher’s model
Figure: evolution in (a) asexual and (b) sexual
populations. The mutations A, B and C are all
advantageous. In the asexual population, an AB
individual can arise only if the B mutation arises in
an individual that already has an A mutation (or vice
versa.) In the sexual population, the AB individual
can be more easily formed by breeding of a B
mutation-bearing individual with an A mutationbearing individual. (c) If favorable mutations are
rare, each will have been fixed before the next
mutation arises, and sexual populations will not
evolve more rapidly.
Role of sexual reproduction in complex genome maintenance?
BENEFICIAL NEUTRAL
DELETERIOUS
GENOME
NATURAL
SELECTION
Costs and benefits
favourable
mutations
haploid
diploid
sexual
asexual
deleterious
mutations
parasitic
sequences
Evolution of sex chromosomes
- many different sex-determining systems in plants and animals with separate
sexes.
- in some species, sex is determined by environmental factors (control expression
of genes leading to male or female development)
- other species have evolved genetic systems involving specialized sex
chromosomes.
- sex chromosomes have arisen independently in many animal groups. Also found
rarely in plants.
It looks like sex chromosomes were once homologs (a pair of equivalent
autosomes—the non-sex chromosomes) that evolved different morphology
and gene content because they lost their ability to recombine. Suppression
of recombination is thought to start around the sex-determining region, but
may eventually affect much of the sex chromosomes. In the absence of
recombination, the two chromosomes of a pair evolve separately and one of
the often deteriorates. Unequal genetic load must be then compensated by
some mechanism.
Faust’s exercise of
making males and females
from hermaphrodites
start with a hermaphrodite with 4 chromosomal pairs
make two sexes with sex chromosomes in a few steps
Extreme variability in regulation of sex determination
each taxon – different solution
>350 MYA
MAMMALIA
Mus
AMPHIBIA
Xenopus
PISCES
Danio
Why is it evolving so fast?
>400 MYA
CHORDATA
>600 MYA
DEUTEROSTOMES
ECHINODERMATA
COELOMATES
PROTOSTOMES
Strongylocentrotus
ARTHROPODA
Drosophila
NEMATODA
Caenorhabditis
EUMETAZOA
PSEUDOCOELOMATES
Haplodiploid sex determination
Complementary sex determination
• ploidy determined by fertilization (no sex chromosomes)
• observed by a priest Johann Dzierzon in 1845: virgin queens, which have
not taken a mating flight produce only male progeny = the first description of
sex determination!
• male bee (drone) n=16 chromosomes, develop from unfertilizede ggs
• female (worker or queen) 2n=32 chromosomes
• about 20% of animals use haplodiploid mode – males are parthenogenotes
developing from unfertilized eggs, females develop from fertilized eggs
However, there is one problem:
Inbreeding studies yielded also diploid males!
How do you explain that?
Complementary sex determination
• complementary sex determination locus model
• heterozygosity required for a female (2 different sex determining alleles)
NORMAL BREEDING
heterozygosity = females
female
male
hemizygosity = males
unfertilized eggs fertilized eggs
male male
female
female
Complementary sex determination
homozygosity is lethal to
males because diploid
males are eaten by
workers soon after they
hatch from eggs, leaving
empty cells behind
INBREEDING
female
male
heterozygosity = females
hemizygosity = males
homozygosity = males
unfertilized eggs fertilized eggs
male male
male
female
Is there any molecular evidence for this wild story?
• yes, indeed. Complementary sex determiner gene (csd)
• csd is a potential splicing factor existing in at least 15 allelic variants
• csd inactivation causes switch to males,
•csd targets fem gene, fem is spliced differently in female and male cells.
http://www.nature.com/scitable/topicpage/sex-determination-in-honeybees-2591764
Sex determination involving sex chromosomes
XX/X0 sex determination
- females have two copies of the sex chromosome (XX)
- males have only one (X0). The 0 denotes the absence of a second sex chromosome.
- found in numerous insects (grasshoppers, crickets, and cockroaches) and other invertebrates.
- C. elegans: male with one sex chromosome (X0); hermaphrodite with a pair of chromosomes (XX).
XX/XY sex chromosomes
- females have two of the same kind of sex chromosome (XX)
- males have two distinct sex chromosomes (XY).
- found in most mammals and insects (Drosophila).
- mammals have a SRY gene on the Y chromosome that determines maleness
- fruit fly use the presence of two X chromosomes to determine femaleness.
ZW sex chromosomes
- ZW sex-determination system is reversed compared to the XY system
- females have two different kinds of chromosomes (ZW)
- males have two of the same kind of chromosomes (ZZ).
-found in birds and some insects (Lepidoptera = butterflies) and other organisms.
Genes in the ZW region in birds are autosomal in mammals, and vice-versa; therefore, it is theorized that the
ZW and XY couples come from different chromosomes of the common ancestor.
A paper published in 2004 (Frank Grützner et al, Nature; DOI:10.1038/nature03021) suggests that the two
systems may be related. According to the paper, platypuses have a ten-chromosome–based system, where
the chromosomes form a multivalent chain in male meiosis, segregating into XXXXX-sperm and YYYYYsperm, with XY-equivalent chromosomes at one end of this chain and the ZW-equivalent chromosomes at the
other end.
Different strategies to compensate unequal genetic load
XX/XY
upregulation of expression in males
XX/XY
silencing of one chromosome in females
XX/X0
reducing expression of both chr. in females
Straub 2007
Different strategies to compensate unequal genetic load
Straub 2007
XX/XO
Caenorhabditis elegans
Stothard 2003
• reducing expression of both chromosomes in females
• X:A ratio determines sex and dosage compensation
• dosage compensation complex (DCC) - at least poly10 peptides
XX/XO
Caenorhabditis elegans
Stothard 2003
• condensin complexes function during mitosis and meiosis for DNA
compaction and sister chromatid resolution
• DCC recruited to specific binding sites on chromosome X
• mechanism of the actual 50% down-regulation is not clear.
XX/XY
Drosophila melanogaster
• upregulation of expression in males (increased expression from the
X chromosome
Lucchesi 2005
• identification of the dosage
compensation complex – MSL
• dosage compensation involves
chromatin modification: H4K16
• H4K16 is unique among acetylation
marks. In yeasts, it plays a role in
maintaining boundary between
silent and active chromatin
MSL - male-specific lethal
XX/XY
Drosophila melanogaster
RNA
Amrein 2000
HAT activity
Homo sapiens/Mus musculus
XX/XY
- dosage compensation by inactivating one X in female cells
4 steps
Counting
Choice
Initiation
Maintenance
- if more than one, choose to inactivate, so one remains active
- random vs. non-random
- initiation and propagation of chromosome-wide silencing
- throughout subsequent cell division
2 types of X-inactivation
XCI
Xi
Xa
Xic
Xce
= X chromosome inactivation
= inactive X
= active X
= X inactivation center
= X-controlling element (Xist/Tsix)
Imprinted X-inactivation
- in early embryos, extraembryonic lineage (trophoblast and primitive endoderm)
Random X-inactivation
-in the epiblast, completed by 5.5.-6.5 dpc
Meiotic Sex Chromosome Inactivation
Thorvaldsen 2006
X-inactivation and reactivation
escape in PGCs
MSCI
imprinted
imprinted
imprinted
random
escape
Turner 2007
Meiotic Sex-Chromosome Inactivation
PMSC – post-meiotic silencing complex
Turner 2007
Meiotic silencing of unsynapsed chomatin
Thorvaldsen 2006
X-inactivation and reactivation
escape in PGCs
imprinted
imprinted
imprinted
random
escape
X-inactivation during preimplantation development
Imprinted X-inactivation
- inactive X inherited or de novo silencing after fertilization?
- pre-inactivation hypothesis
- sex chromosome inactivation during spermatogenesis
- XY body in spermatocytes, MSCI (meiotic sex chr. inact.)
- MSCI not fully understood, different from XCI (Xist independent,
specific chromatin modifications including histone variant H2AX)
- staining of 2-cell embryos indicate lack of active transcription on
the paternal X
- some data support reversion into the active state after meiosis
- “de novo” model
- Xp active at fertilization, silenced later
- staining of 2-cell embryos showing biallelic expression
- Xist dependent (Xist expressed at the 2-cell stage)
Xist and Tsix
Avner 2001
Xist and Tsix
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
Figure 1. Mouse and Human Xic/XIC and Xist/XIST. A. area surrounding the XIST/Xist gene on human and
mouse X-chromosomes. Human domain is inverted from mouse relative to telomere. The identification of a
human Tsx homolog is unclear. B. Comparison of the mouse Xist and human XIST genes. * = alternative
splicing sites. Mouse has a second promoter that has not been found in other Xist/XIST genes analysed to
date. Extensive alternative splicing of the human gene has been described yielding isoforms that lack exon 4,
half of exon6, exon7 or include the last two introns.
Xist
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
Longest Xist 17.9 kb. Longest XIST 19.3 kb.
Mouse and human Xist/XIST show 49% sequence identity which is lower than 5' &
3' UTR regions but slightly higher than introns.
Several short stretches of high homology and six repeated elements A-F.
No open reading frame. Must operate as polyadenylated RNA.
Xist
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
Tsix
-two promoters and two polyadenylation sites. no significant open reading
frames.
- Tsix transcripts of up to 4 kb can be produced by splicing.
- Tsix is not the counting element (Males lacking Tsix do not inactivate).
- Tsix RNA is antisense to Xist and reduces its steady-state level while
subsequently promotes Xa choice by increasing the affinity of the cis-linked
counting element for blocking factor.
- the spliced form of Tsix RNA contains only 2 kb of overlap with the mature Xist
transcript. This overlap occurs within a domain of Xist that is critical for silencing
activity
- antisense transcription throught the Xist sequence is important (Deletion
mutants lacking the overlap)
Avner 2001
Tsix
X-inactivation during preimplantation development
Imprinted X-inactivation
- paternal Xist expression activated at the 2-cell stage
- Xist silencing of the maternal X (Xm) is unclear
- Tsix (maternal) is detected first at the 8-cell stage
- Xist accumulates on the Xp (initiation event) from the 4-cell on
- initial chromatin changes found during the 8-32-cell stages
- hypoacetylation H3K9
- hypomethylation H3K4
- EED/EZH2 enrichment mediates H3K27 methylation on the Xi
- initiation vs. maintenance changes unknown
- gradient of silencing from the Xic suggests that silencing is
progressive, mediated by Xist RNA spreading
- ICM cells reverse imprinted XCI, trophectoderm cells maintain it
X-inactivation during preimplantation development
Random X-inactivation
- paternal Xist silencing reversed after early blastocyst
- Xist dispersed or absent
- no EED/EZH2 association,
- random X-inactivation initiates during implantation and is
complete around day 6.5 dpc
- initiation is characterized by downregulation of Tsix and
upregulation of Xist
- Xist coats the Xi in cis
- chromatin modifications
- DNA methylation is a late step
- once established, the Xi is clonally propagated such that females
are functionally mosaic for X-linked traits.
- epigenetic modification can later maintain Xi repression in a Xistindependent manner
Thorvaldsen 2006
Xist and Tsix
XCI
Xi
Xa
Xic
Xce
= X chromosome inactivation
= inactive X
= active X
= X inactivation center
= X-controlling element (Xist/Tsix)
Avner 2001
doesn’t seem
to be the case
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
XCI in differentiating female ES cells
MacroH2A is recruited to the Xi by Xist.
followed by exclusion of H2ABbd variant from Xi. (formation of Barr body).
Xist/XIST espression is not necessary to continue Xi after establishment.
DNA methylation appears to be extremely important for the stability and
maintenance of gene silencing on Xi.
DNA methylation concerns promoter regions, overall is the inactive X
hypomethylated!
http://bioweb.wku.edu/courses/biol566/L9XchromSilencing.html
XCI in differentiating female ES cells
Turner syndrome - 45, X or 46, X, abn X
• fairly common (10% od spontaneous abortions)
• 1 of 40 develops to birth, then the phenotypic effects are relatively mild because
each cell has a single functioning X chromosome like those of XX females.
• phenotypic female with gonadal dysgenesis and sexual immaturity, have primary
amenorrhea (failure to menstruate), infertility, short stature, webbed neck,
increased carrying angle at the elbow, cardiovascular and renal abnormalities
• 45,X in more than half the patients
Number of Barr bodies = zero.
Incidence: 1 of 2500 female births
Why does Turner syndrome occur at all, since only one X chromosome is
normally active?
There are two active X chromosomes during ovarian development, and certain
genes appear to need to be active for normal ovarian function. Turner syndrome
oocytes virtually gone by the age of 2 years
Klinefelter syndrome - 47, XXY(48, XXXY)
• males (Y chromosome).
• the phenotypic effects of the extra X chromosomes are mild because, the
extra Xs are inactivated and converted into Barr bodies
• male with small testes, hyalinized testicular tubules, and azoospermia
(failure to produce normal amounts of sperm), resulting in infertility and
variable signs of hypogonadism, social pathologies, somewhat reduced IQ,
postpubertal testicular failure
• may have additional X chromosomes, if so, more likely to be mentally
retarded
• demonstration in humans that sex is determined by presence or absence
of Y chromosome, rather than number of X chromosomes
Number of Barr bodies = extra X’s inactivated
Incidence: 1 of 1000 male births
XYY syndrome
- found as 47,XYY, or 48,XXYY
47,XYY
- occurs 1/1000 in male live births
- occurs 4-20 per 1000 inmates
48,XXYY
- incidence 1/20-40 000
- 50 times higher in prison inmates than in newborn population
XX males
- incidence 1 in 20,000
- have X-Y interchange
- Sry transgenic mice, XX become male
XXX, XXXX, XXXXX females
- mild phenotypic effects because in each cell all the extra X
chromosomes are inactivated.
- number of Barr bodies = number of X chromosomes minus one.
IMPRINTING
Discovery of Imprinted Genes
• experimental manipulation of mouse embryos in the early 1980's showed that
normal development requires the contribution of both the maternal and paternal
genomes.
• gynogenetic embryos (two female genomes) show relatively normal
embryonic development, but poor placental development.
• androgenetic embryos (two male genomes) show very poor embryonic
development but normal placental development.
• it is now known that there are around 100 imprinted genes in humans and
mice, many of which are involved in embryonic and placental growth and
development
• the gynogenetic embryos have twice the normal level of maternally expressed
genes, and completely lack expression of paternally expressed genes, whereas
the reverse is true for androgenetic embryos.
• no naturally cases of parthenogenesis exist in mammals (Jesus does’nt count!)
• manipulation of a paternal methylation imprint controlling the Igf2 locus allowed
the creation of rare individual mice with two maternal sets of chromosomes (not
a true parthenogenote).
http://atlasgeneticsoncology.org/Deep/GenomImprintID20032.html
Mouse germ cell pronuclear transplant experiments convincingly demonstrate a
different agenda for sperm- versus egg-derived nuclear genomes during
development. Development in the absence of a sperm-derived genome (middle
column) shows fairly good development of the embryo proper but failed
development of the trophoblast lineage. Development in the absence of an eggderived genome (right column) shows failed development of the embryo proper
but exuberant trophoblast growth.
Imprinting is a cause of phenotypes in uniparental disomies
• In 1980 Engel introduced the concept of uniparental disomy (UPD).
• Uniparental disomy (UPD) arises when an individual inherits two copies of a
chromosome pair from one parent and no copy from the other parent.
• In the rare circumstance of UPD a baby may have two copies of one of his/ her
mother’s chromosome and no copies of that chromosome from his/ her father.
This is called maternal UPD. Paternal UPD is when a child inherits two copies of
a specific chromosome from his/ her father and no copies of that chromosome
from his/ her mother.
•This abnormality in inheritance may lead to health concerns in a child.
• UPD can result in rare recessive disorders, or developmental problems due to
the effects of imprinting. UPD may also occur with no apparent impact on the
health and development of and individual.
Prader-Willi syndrome, Angelman syndrome, Beckwith-Wiedemann syndrome
Described in detail in Buiting 2010
Described in detail in Choufani 2010
http://www.mgu.har.mrc.ac.uk/research/imprinting/imprin-viewmaps.html
Maps of imprinted genes
General features of imprinted genes
• typically clustered, clusters may contain monoallelic expression of genes from
each parent.
• clusters contain imprint control regions and a non-coding RNA is often found
associated with it (H19, Air …)
• ICRs show parent-of-origin dependent epigenetic modifications (methylation)
• many related to growth control (battle of the sexes hypothesis)
• it has been reported that imprinted genes tend to have smaller introns.
• some genes imprinted only in neural tissues
Robertson 2005
Mouse distal 7 imprinted region
H19 - a noncoding RNA!!
CTCF
http://www.ncbi.nlm.nih.gov/entrez/dispomim.cgi?id=604167
• CCCTC binding factor with 11 zinc fingers
• highly conserved in vertebrates (93% identity human-avian)
• binds to regulatory sequences in numerous loci, including H19/IGF2
• binding is methylation sensitive, protects DNA from methylation
• insulator - regulates access of enhancers/separates functional domains
• boundary element - blocks spread of heterochromatin
Fedoriw 2004
Binding of CTCF is essential for
proper H19/IGF2 imprinting
H19 is a non-coding RNA
• ~2.3 kb long, maternally expressed
• integrity of elements controlling H19 transcription essential for Igf2 imprinting
• H19 RNA is not essential for Igf2 imprinting
• ectopic H19 overexpression can affect viability but targeted deletion makes no
obvious phenotype (but expression of other is genes affected)
• H19 could be a primary miRNA precursor
For recent data se Gabory et al., Bioessays 2010
Reciprocal imprinting, ICR and non-coding RNAs is a common theme
Lucifero 2004
Variable timing of maternal imprinted marks
De novo DNMT3a/b, DNMT3L
Two modes of imprinting
Insulator model
ncRNA model
Air ncRNA expression involves in silencing of upstrean genes
IMPRINTING DEFECTS IN ARTs
X-inactivation and imprinting evolution
Pauler 2007