Dr. Yaniv Erlich
advertisement

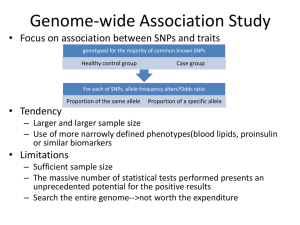
Exome sequencing and disease-network analysis Yaniv Erlich Whitehead Institute for Biomedical Research 4/20/11 DNA Sudoku Yaniv Erlich Two projects with neurological diseases 1. Joubert syndrome in Ashkenazi Jews 2. Hereditary Spastic Paraparesis in Palestinians yaniv@wi.mit.edu Our approach Intro. Results Conclusion What is Joubert Syndrome? • Cerebello-oculo-renal phenotype: - Hypoplasia of the cerabellar vermis - Pyschomotor retardation and hyptonia - Extra digits in upper and lower limbs (sometimes) - Lazy eye - Renal insufficiency • Molar Tooth Sign in MRI: Normal Joubert Thick and elongated superior cerebellar peduncles Large Interpeduncular fossa Cerebellum Parisi, 2007 Exome sequencing and disease network yaniv@wi.mit.edu Our approach Intro. Results Conclusion The cases • Dor-Yeshorim identifies 13 cases in Jewish Ashkenazi families • 8 families, where 3 are part of the same clan. • Autosomal recessive pattern • Founder effect Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion The starting point • 9 genes are known to cause Joubert syndrome in other populations • Sequencing those genes revealed normal results • Social implication… • Using autozygoustity mapping Hadassah found a strong signal from the centromeric region of chromosome 11. Chr 11 Hundreds of exons… Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Finding the mutation – our approach • Sequence the entire exome of a healthy mother and an affected daughter. • Finding mutations that are: - Heterozygous in the mother - Homozygous in the child - Not in dbSNP - Causing a change in the coding region • Two lanes 36x2 per specimen • Hadassah sequenced the exons in the region according to a prioritized list of candidate exons. Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Technical details Step Tool Comments Sequencing Illumina Mother: ~67,400,000 single end-reads Child: ~73,600,000 single end-reads Alignment Bowtie - Default parameters -> poor results. (only 1% of reads were mapped). - Iterative mapping Total unique mapper: Mother – 51 million (76%) Child – 54 million (74%) SNP calling SOAPsnp Only regions with x5 coverage or more Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Conclusion Results Analysis of variations SNPs in: # Mother 49,515 Child 48,142 That are shard… 23,986 And are heterozygous in mother and homozygous in child… And not in dbSNP And are not synonymous And are in the mapped locus Exome sequencing and disease network 2,541 105 39 1 yaniv@wi.mit.edu Intro. Our approach Results Conclusion And the winner is… TMEM216 Chromosome 11 Amino-acid change: Arg -> Leu (CGC>CTC) Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Who are you TMEM216? • Transmembrane protein • 88 amino-acid • Not a single paper on that gene • Conservation analysis: Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Additional lines of evidence • PolyPhen predicated as damaging •Hadassah found the same mutation (double blind) - Found in all 13 cases and parents found as carriers. From: Elpeleg Orly Sent: Sun 12/6/2009 11:56 PM To: Erlich, Yaniv Subject: RE: Preliminary analysis BINGO ________________________________________ From: Erlich, Yaniv [erlich@cshl.edu] Sent: Sun 12/6/2009 23:32 To: Elpeleg Orly Subject: Preliminary analysis Hi Orly, The only potential homozygous SNP mutation we found on chr11 between 59.5M-62M that is not in dbSNP and has an affect on the protein (missense, nonsense, splice) is TMEM216 Arg12->Leu (chr11:60918013). Are we right? Thanks, Yaniv Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Summary • We found that TMEM216 is the causative mutation of Joubert syndrome in Ashkenazi Jews. • The project took 2 months end-to-end in our side. • We used only 4 lanes of Illumina GAII with paired-end 36nt reads. • Israel Ministry of Health added the mutation to their test panel. • Carrier rate in Ashkenazi Jews is 1:92 Exome sequencing and disease network yaniv@wi.mit.edu Intro. Our approach Results Conclusion Two projects with neurological diseases 1. Joubert syndrome in Ashkenazi Jews 2. Hereditary Spastic Paraparesis in Palestinians yaniv@wi.mit.edu Intro. The problem Our approach Results Conclusion Hereditary Spastic Paraperesis -A single Palestinian family: - - 3 brothers suffers from progressive weakness of the legs and abnormal gait - Phenotype is HSP – degradation of the pyramidal tract. - 20 genes have been documented 4/20/11 Exome sequencing and disease network (Weber J,2003) Yaniv Erlich Intro. Our approach The problem Results Conclusion Thoughts on the Joubert rejection process Currently, the rejection process is based on two classes of arguments: 1. Genetic arguments 2. Loss of function arguments SNPs in: # Mother 49,515 Child 48,142 That are shard… Genetic arguments Loss-offunction arguments 23,986 And are heterozygous in mother and homozygous in child… And not in dbSNP 2,541 105 And are not synonymous And are in the mapped locus 39 1 Genetic arguments have weaker power with smaller families We need a new class of arguments Exome sequencing and disease network yaniv@wi.mit.edu Intro. The problem Our approach Results Conclusion Disease-network analysis Idea: Genes with a similar phenotype have a similar biological signature Zippi Brownstein, 2009 Method: Stratify variations according to their similarity to known disease genes. Exome sequencing and disease network yaniv@wi.mit.edu Intro. The problem Our approach Results Conclusion Data gathering stage Whole exome seq. with Illumina Whole genome genotyping with Affy 250K array 4/20/11 Exome sequencing and disease network Yaniv Erlich Intro. The problem Our approach Results Conclusion Results • 20 known genes that cause HSP were found intact • None of the 40,000 disease causing mutations in HGMD were found in the exome sequencing data. • X-linked mutation was ruled out. • Genotyping identified 4 regions of homozygosity: Conclusion: it is a new gene 4/20/11 Exome sequencing and disease network Yaniv Erlich Intro. The problem Our approach Results Conclusion Exclusion process Total Genes Position Sequenced Genes Position Now what? 4/20/11 Exome sequencing and disease network Yaniv Erlich Intro. The problem Our approach Results Conclusion Disease network analysis rank KIF1A as the top candidate 4/20/11 Exome sequencing and disease network Yaniv Erlich Intro. The problem Our approach Results Conclusion Validation by loss of function analysis Total Genes 4/20/11 Position Exome sequencing and disease network Sequenced Genes Position Yaniv Erlich Intro. The problem Our approach Results Conclusion Supporting evidence • Sanger sequencing confirmed the mutation in third affected child. 4 healthy brothers were not homozygous for the mutation • KIF1A is a kinesin. Phenotype is neuronal. • The region of KIF1A was suspected to cause HSP by a previous study with multiple Algerian family. • Conservation suggests a functional domain in KIF1A 4/20/11 Exome sequencing and disease network Yaniv Erlich Intro. The problem Our approach Results Conclusion Summary • Identifying a new gene for spastic paraperesis. • Using only a single family and a new class of arguments •The rate of the mutation is 1:200 in Palestinians. Exome sequencing and disease network yaniv@wi.mit.edu Erlich Lab Whitehead Institute Positions available (yaniv@wi.mit.edu) yaniv@wi.mit.edu