Genome-wide Association Study
advertisement
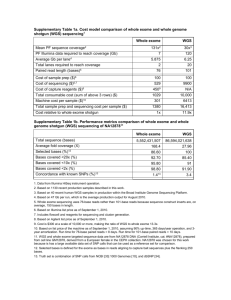
Genome-wide Association Study • Focus on association between SNPs and traits genotyped for the majority of common known SNPs Healthy control group Case group For each of SNPs, allele frequency alters?Odds ratio • Tendency Proportion of the same allele Proportion of a specific allele – Larger and larger sample size – Use of more narrowly defined phenotypes(blood lipids, proinsulin or similar biomarkers • Limitations – Sufficient sample size – The massive number of statistical tests performed presents an unprecedented potential for the positive results – Search the entire genome-->not worth the expenditure Advantage of Exome Sequecing • Whole genome sequencing – Redundant raw data(6 Gb in each human diploid genome ) • Exome sequecing(targeted exome capture) – Exons are short and 180,000 exons constitute 1% of the human genome – The goal is to identify the functional variation that is responsible for both mendelian and common diseases Significance • Exome sequencing can be used to identify causal variants of rare disorders • The first reported study that used exome sequencing as an approach to identify an unknown causal gene for a rare mendelian disorder The Shendure Lab • Next-generation human genetics – A multiplex approach to genome sequencing – Targeted sequence enrichment • Protocols relying on molecular inversion probe • Hybrid capture – Novel analytical strategies to identify the genetic basis of Mendelian disorders by exome sequecing • Autosomal recessive disorders such as Miller syndrome • Autosomal dominant disorders such as Kabuki syndrome Hapmap project • Focuse on common SNPs(at least 1% of the population) • Samples: 4 populations – (30*3 YRI, 30*3 CEU, 45 JPT, 45 CHB) • Data: – SNP frequencies, genotypes Work flow DNA samples, targeted capture and massively parallel sequencing Read mapping and variant analysis Direct identification of the causal gene for FSS Target enrichment Methods a. PCR-based approach b. Molecular inversion probe(MIP)based approach c. Hybrid capture-based approach Mamanova et al. Nat Method 7(2):112-118 Mamanova et al. Nat Method 7(2):112-118 Figure. ①Probe list of array2 ② Probe list of array1 ③Exome on 1-22, X and Y chromosomes Work flow DNA samples, targeted capture and massively parallel sequencing Read mapping and variant analysis Direct identification of the causal gene for FSS Coming… Comparison of sequence calls to array genotypes, dbSNP and whole genome sequencing Direct identification of the causal gene for FSS Method Genomic DNA samples Oligonucleotides and adaptors Shotgun library construction Design of exon capture array Targeted capture by hybridization of DNA microarrays Sequencing Read mapping Target Masking Variant calling Comparison of sequence calls to array genotypes, dbSNP and whole genome sequencing Variant annotation Calculation of genome-wide estimates Method Method 2:MIP and resequencing Method 3: Whole genome sequencing Method 4: Figure. Table of cSNPs of 8 HapMap individuals Figure. Table of Splice Site Variants of 8 HapMap individuals Figure. Table of Coding Indels of 8 HapMap individuals Figure. Table of coverage of 8 HapMap individuals and 4 FSS individual Figure. Intervals that were exclued…. Figure. ①Probe list of array2 ② Probe list of array1 ③Exome on 1-22, X and Y chromosomes YRI: Nigeria - Yoruba people of Ibadan CHB: China - Beijing JPT: Japan - Tokyo CEU: Centre d'Etude du Polymorphisme Humain (CEPH) Eur: European–American ancestry About mendelian disease Traditional situation Current situation Considerations • • • • Causal genes may be shared by case group. Control group may not contain that mutation. Common mutation may not be causal. Causal mutation should cause animo acid change. Result Further application • Typical single gene disorder. • Disorder caused by single but not uniform gene. • Multiple gene disorder. • Complex disease. • Cancer.