Finding The Lost Treasure of Sequencing data
advertisement

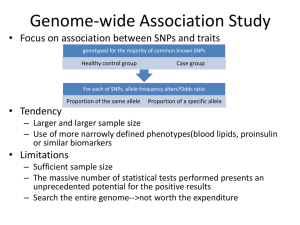
Vanderbilt Center for Quantitative Sciences Summer Institute Sequencing Analysis (DNA) Yan Guo Alignment ATCGGGAATGCCGTTAACGGTTGGCGT Reference genome Human genome is about 3 billion base pair (3,000,000,000)in length. If read is 100 bp long, what is the probability of unique alignment? 1/(4x4x4…4) =1/4100 =1/1.60694E+60 Alignment Tools • BWA http://bio-bwa.sourceforge.net/ • Bowtie http://bowtiebio.sourceforge.net/index.shtml Doing accurate alignment for a 30 million reads will take 30 million x 3billion time units. Both are based on Borrows-Wheeler Algorithm Alignment Results – Bam files • SAM – uncompressed • Bam – compressed • http://samtools.github.io/htsspecs/SAMv1.pdf • Sort and index before performing analysis • Don’t forget to perform QC on alignment How to call SNPs http://www.broadinstitute.org/igv/ Local Realignment Recalibration Why do we need realignment and recalibration for DNA but not RNA? SNP calling • GATK https://www.broadinstitute.org/gatk/ • Varscan http://varscan.sourceforge.net/ VCF files Annotation using ANNOVAR http://www.openbioinformatics.org/annovar/ Somatic Mutation • Different from SNP (not germline) • Both tumor and normal samples are needed to accurately define a somatic mutation • Tumor sample is almost never 100% tumor Somatic mutation callers • MuTect http://www.broadinstitute.org/cancer/cga/m utect • Varscan http://varscan.sourceforge.net/ Quality Control on SNPs • Number of Novel Non-synonymous SNP ~ 100 – 200 • Transition / transversion ratio • Heterozygous / non reference homozygous ratio • Heterozygous consistency • Strand Bias • Cycle Bias Ti/Tv ratio Heterozygous / non reference homozygous ratio Ti/Tv ratio by race and regions Heterozygous / non reference homozygous ratio by race and regions Heterozygous Genotype Consistency Strand Bias Table 1 . Strand bias examples from real data Chr Pos depth a1 b2 c3 d4 6 32975014 21 5 5 10 1 1 81967962 38 20 11 7 0 12 10215654 31 15 7 0 9 1. Forward strand reference allele 2. Forward strand non reference allele 3. Reverse strand reference allele 4. Reverse strand non reference allele Forward Strand Genotype Reverse Strand Genotype Heterzygous Homozygous Heterzygous Homozygous Heterzygous Homozygous Cycle Bias Pooled Analysis • Pool samples together without barcode • Save money • Can only be used to evaluate allele frequency Pooled Analysis - Conclusion Advanced Data Mining The known and unknown of sequencing data The known and unknown of sequencing data Known Unknown The known and unknown of sequencing data Known Known Unknown Unkown Unkown Known – Things we always know that Sequencing data can do SNV, mutation CNV Xie et al. BMC Bioinformatics 2009 Structural Variants Alkan et al. Nature Review Genetics, 2011 Known Unknown – Other information we found that sequencing data contain Known Known Unknown Unkown Unkown How is additional data mining possible? • Data mining is possible because capture techniques are not perfect. Capture Efficiency of The Three Major Capture Kits Potential Functions of Intron and Intergenic ENCODE suggested that over 80% human genome maybe functional. Majority of the GWAS SNPs are not in coding regions (706 exon, 3986 intron, 3323 intergenic) Coverage of the Unintended Regions • The coverage don’t just drop off suddenly after the capture region end. • Capture region example: chr1 1000 1500 1000 1500 1000 1500 Reads Aligned to Non Target Regions Can Be Used to Detect SNPs • Tibetan exome study : Through exome sequencing of 50 Tibetan subjects, 2 intron SNPs were identified to be associated with high altitude. (Yi, et al. Science 2010) • Non capture region study: Non capture region’s reads were studied to show they can infer reliable SNPs. (Guo, et al BMC Genomics) Known unknown - Mitochondria However, mitochondria is only 16569 BP Assumptions: 40 mil reads 100BP long read Dealing with nuMTs Alignment Results Extract mitochondria from exome sequencing Tools: • Picardi et al. Nature Methods 2012 • Guo et al. Bioinformatics, 2013 (MitoSeek) Diagnosis: • Dinwiddie et al. Genmics 2013 • Nemeth et al, Brain 2013 Virus • Virus sequences can be captured through high throughput sequencing of human samples • HBV in liver cancer samples (Sung, et al. Nature Genetics, 2012) (Jiang, et al. Genome Research, 2012) • HPV in head and neck cancer (Chen, et al. Bioinformatics, 2012) HPV AlignmentExample Tools for Detecting Virus from Sequencing data • PathSeq (Kostic, et al. Nature, 2011 Biotechnology) • VirusSeq (Chen, et al. Bioinformatics, 2012) • ViralFusionSeq (Li, et al. Bioinformatics, 2012) • VirusFinder (Wang, et al. PlOS ONE, 2013) The Data Mining Ideas applied to RNA • RNAseq has been used a replacement of microarray. • Other application of RNAseq include dection of alternative splicing, and fusion genes. • Additional data mining opportunities also available for RNAseq data SNV and Indel • Difficulty due to high false positive rate • RNAMapper (Miller, et al. Genome Research, 2013) • SNVQ (Duitama, et al. (BMC Genomics, 2013) • FX (Hong, et al. Bioinformatics, 2012) • OSA (Hu, et al. Binformatics, 2012) Microsatellite instability Examples: • Yoon, et al. Genome Research 2013 • Zheng, et al. BMC Genomics, 2013 RNA Editing and Allele-specific expression RNA editing tools and database • DARNED, REDidb, dbRES, RADAR Allele-specific expression • asSeq (Sun, et al. Biometrics, 2012) • AlleleSeq (Rozowsky, et al. Molecular Systems Biology, 2011) Exogenous RNA • Virus (Same as DNA) • Food RNA (you are what you eat) Wang, et al. PLOS ONE, 2012 nonCoding RNA Unknown Unknown Exome Samuels, et al. Trends in Genetics, 2013 RNAseq Quality Control Quality Guo et al. Briefings in Bioinformatics, 2013 Quantity