Lecture 8 LC710- 1st + 2nd hr
advertisement

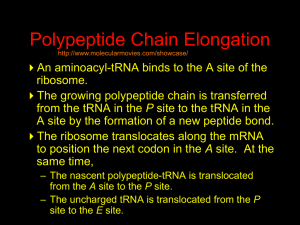
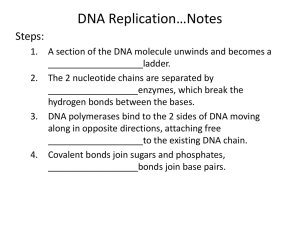
Hybridization of Nucleic Acids DNA1 DNA2 RNA Probe Northern hybridization Southern hybridization Juang RH (2004) BCbasics Preparation of Traditional Nucleic Acid Probe Amino acid sequence GLY-ASP-GLU-SER-SER-VAL-LEU----GGG-GAC-GAG-TCC-TCC-GTT-CTC--- * * * * Nucleic acid sequence The nucleic acid sequence is Deduced from amino acid sequence Chemical synthesis * * * * Codon degeneracy Synthesizing oligonucleotide PROBE: GGGGACGAGTCCTCCGTTCT Juang RH (2004) BCbasics Probe is labeled with radioactive 32P 32 P GG GGACG AG Hybridization GG T TC CT CC GTTCT CA TGCTC C C C G AGG G TC A A G AGGCAA C C T A DNA denaturation Target gene Single colony Lysed Juang RH (2004) BCbasics Colony Is Screened by Hybridization with Probe Colony hybridization Transferring … Collect filter paper Dissolve cell Autoradiography DNA denatured Add probe Juang RH (2004) BCbasics Cover with filter paper Biochip Based on Hybridization Sample DNA Complementary DNA hybridize Biochip Each spot contains known DNA Signal appears Schena (2000) Microarray Biochip Technology, p. A31 Juang RH (2004) BCbasics The Genetic Code Initiation and termination Codons – Initiation codon: AUG – Termination codons: UAA, UAG, UGA Degeneracy: partial and complete Ordered Nearly Universal (exceptions: mitochondria and some protozoa) Key Points Each of the 20 amino acids in proteins is specified by one or more nucleotide triplets in mRNA. (20 amino acids refers to what is attached to the tRNAs!) Of the 64 possible triplets, given the four bases in mRNA, 61 specify amino acids and 3 signal chain termination. (have no tRNAs!) Key Points The code is nonoverlapping, with each nucleotide part of a single codon, degenerate, with most amino acids specified by two to four codons, and ordered, with similar amino acids specified by related codons. The genetic code is nearly universal; with minor exceptions, the 64 triplets have the same meaning in all organisms. (this is funny) Do all cells/animals make the same Repertoire of tRNAs? The Genetic Code The Wobble Hypothesis: Base-Pairing Involving the Third Base of the Codon is Less Stringent. Base-Pairing with Inosine at the Wobble Position In molecular biology, a wobble base pair is a non-WatsonCrick base pairing between two nucleotides in RNA molecules. The four main wobble base pairs are guanine-uracil, inosine-uracil, inosine-adenine, and inosinecytosine (G-U, I-U, I-A and I-C). The thermodynamic stability of a wobble base pair is comparable to that of a Watson-Crick base pair. Wobble base pairs are fundamental in RNA secondary structure and are critical for the proper translation of the genetic code. Suppressor Mutations Some mutations in tRNA genes alter the anticodons and therefore the codons recognized by the mutant tRNAs. These mutations were initially detected as suppressor mutations that suppressed the effects of other mutations. Example: tRNA mutations that suppress amber mutations (UAG chain-termination mutations) in the coding sequence of genes. Making a (UAG) Mutation Translation of an amber (UAG) Mutation in the Absence of a Suppressor tRNA Translation of an amber Mutation in the Presence of a Suppressor tRNA Note it is amber su3…why????????? Translation of an amber Mutation in the Presence of a Suppressor tRNA If there was a single tRNATyr gene, then could one have a amber supressor of it? Fig1 Oligonucleotide synthesis is carried out by a stepwise addition of nucleotide residues t o the 5'-terminus of the growing chain until the desired sequence is assembled. Each addition is referred to as a synthetic cycle (Scheme 6) and consists of four chemical reactions: * Step 1 - De-blocking (detritylation): The DMT group is removed with a solution of an acid, such as T CA or Dichloroacetic acid (DCA), in an inert solvent (dichloromethane or t oluene) and washed out , resulting in a free 5' hydroxyl group on the first base. * Step 2 - Coupling: A nucleoside phosphoramidite (or a mixt ure of several phosphoramidites) is activated by an acidic azole catalyst, tetrazole, 2-ethylthiotetrazole, 2-bezylthiotetrazole, 4,5-dicyanoimidazole, or a number of similar compounds. This mixture is brought in contact with the starting solid support (first coupling) or oligonucleotide precursor (following couplings) whose 5'-hydroxy group reacts with the activated phosphoramidite moiety of the incoming nucleoside phosphoramidite to form a phosphite triester linkage. This reaction is very rapid and requires, on small scale, about 20 s for its completion. The phosphoramidite coupling is also highly sensitive to the presence of water and is commonly carried out in anhydrous acet onitrile. Unbound reagents and by-products are removed by washing. * Step 3 - Capping: After the completion of the coupling reaction, a small percentage of the solid support-bound 5'-OH groups (0.1 t o 1%) remains unreacted and needs to be permanently blocked from further chain elongation to prevent the formation of oligonucleotides with an internal base deletion commonly referred to as (n-1) shortmers. This is done by acetylation of the unreacted 5'-hydroxy groups using a mixture of acetic anhydride and 1-methylimidazole as a catalyst. Excessreagents are removed by washing. * Step 4 - Oxidation: The newly formed tricoordinated phosphite triester linkage is not natural and is of limited stability under the conditions of oligonucleotide synthesis. The treatment of the support-bound material with iodine and water in the presence of a weak base (pyridine, lutidine, or collidine) oxidizes the phosphite triester into a tetracoordinated phosphate triester, a protected precursor of the naturally occurring phosphate diester internucleosidic linkage. This step can be substituted with a sulfurization step to obtain oligonucleotide phosphorothioates (see below). In the lat ter case, the sulfurization step is carried out prior to capping. New Base Are the proteins produced a pure reflection of the mRNA sequence???? tRNA environment, protein modifications post-translationally Good things to Know RNApol II TATAA CCATGG (Nco I site and Kozak Rule) ATG AGGT….splice GT……………A………polypyrimidine AG PolyA recog sequence AATAAA The Reasons why ATG is a single codon and TGG is a single codon. SELEX yields a functional binding site. A, COS7 cells were transfected in triplicate with either pEBG or increasing concentrations of BENwt (250, 500, and 1000 ng) and either p81TKluc (TK) luciferase reporter or p81TKluc-WT3X (WT3X). The luciferase values are reported as relative luciferase activity normalized to the amount of total protein. -Fold decrease in activity is measured relative to the basal transcriptional activity observed with pEBG empty expression vector alone. Western blot with anti-GST antibody shows dosedependent expression of GST-BEN. B, COS7 cells were transfected in triplicate with either pEBG or BENwt (1000 ng of each) and with either TK, or WT3X or Mut3X (600 ng of each) and the Renilla construct (pRL-TK).