File
advertisement

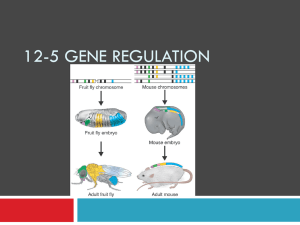
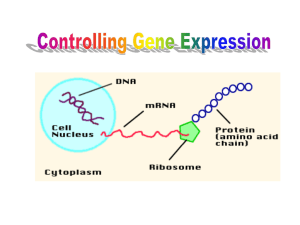
Gene Regulation Is Necessary ~42 000 genes exist that code for proteins in humans, but not all proteins are required By switching genes off when they are not needed, cells can prevent resources from being wasted. The ability to switch genes on an off is naturally selected A typical human cell normally expresses about 3% to 5% of its genes at any given time. Cancer results from genes that do not turn off properly. Cancer cells have lost their ability to regulate mitosis, resulting in uncontrolled cell division. Gene expression in eukaryotes is controlled by a variety of mechanisms that range from those that prevent transcription to those that prevent expression after the protein has been produced. The various mechanisms can be placed into one of these four categories: transcriptional, posttranscriptional, translational, and posttranslational. Transcriptional prevent mRNA from being synthesized Ex: Barr bodies in females are inactive because tightly wound Post-transcriptional Regulate mRNA after it is produced Ex: a single mRNA can code for 3 proteins, depending on which introns are removed Translational Prevent protein synthesis Proteins may bind to regions of the mRNA strand preventing the ribosomes from translating it. Post-translational Prevent the protein from becoming functional Ex: Proteins are often not fully functional after translation. Proinsulin is a precursor to insulin. It needs to be cut into 2 polypeptide chains and have 30 amino acids removed. Prokaryotes Much of our understanding of gene control comes from studies of prokaryotes. Prokaryotes have two levels of gene control. Transcriptional and translational. Operons Operons are groups of genes that function to produce proteins needed by the cell. Prokaryotic cells use operons to regulate genes and their respective proteins. Operons are made up of: Structural Genes – code for the proteins needed. Ex: the proteins needed to breakdown sugar Promoter – are where RNA polymerase binds to the DNA (Lots of A-T base pairs!) Operator – a short sequence of bases between structural genes and a promoter. The lac operon Lactose is a sugar found in milk. If lactose is present, E. coli (the common intestinal bacterium) needs to produce the necessary enzymes to digest it. Three different enzymes are needed. In the diagram genes A, B, and C represent the genes whose products are necessary to digest lactose. In the normal condition, the genes do not function because a repressor protein is active and bound to the DNA preventing transcription. When the repressor protein is bound to the DNA, RNA polymerase cannot bind to the DNA. The protein must be removed before the genes can be transcribed. Lac operon – with repressor (no transcription) Below: Lactose binds with the repressor protein inactivating it. Transcription! Lac operon The lac operon is an example of an inducible operon because the structural genes are normally inactive. They are activated when lactose is present. The trp operon Repressible operons are the opposite of inducible operons. Transcription occurs continuously and the repressor protein must be activated to stop transcription. Tryptophan is an amino acid needed by E. coli and the genes that code for proteins that produce tryptophan are continuously transcribed as shown below. However, if tryptophan is present in the environment, E. coli does not need to synthesize it and the tryptophan-synthesizing genes should be turned off. This occurs when tryptophan binds with the repressor protein, activating it. Unlike the repressor discussed with the lac operon, this repressor will not bind to the DNA unless it is activated by binding with tryptophan. Tryptophan is therefore a co-repressor. The trp operon is an example of a repressible operon because the structural genes are active and are inactivated when tryptophan is present. Negative and Positive Control The trp and lac operons discussed above are examples of negative control because a repressor blocks transcription. In one case (lac operon) the repressor is active and prevents transcription. In the other case (trp) the repressor is inactive and must be activated to prevent transcription. Positive control mechanisms require the presence of an activator protein before RNA polymerase will attach. The activator protein itself must be bound to an inducer molecule before it attaches to mRNA. Genes which code for enzymes necessary for the digestion of maltose are regulated by this mechanism. Maltose acts as the inducer, binding to an activator and then to DNA. The activator bound to DNA stimulates the binding of RNA polymerase. Methylation The attachment of a methyl group to histone proteins can promote or inhibit transcription (by either causing the DNA to unravel from the nucleosome or stay tightly bonded to the nucleosome). If the methyl group is attached directly to DNA, transcription will be inhibited because RNA polymerase cannot attach. Methylation of Cytosine Methylation of a Nucleosome Methylation The amount of DNA methylation varies during a lifetime and is affected by environmental factors. Identical twins will have identical DNA but will have different levels of methylation because they have different experiences. Differences in the expressed genes is why identical twins may not look or act exactly the same. Epigenetics Epigenetics is the study of cellular and physiological traits that are NOT caused by changes in the DNA sequence. This done via chemical modifications (such as methylation, phosphorylation, or acetylation) and are called epigenetic tags. Scientific evidence shows that not only does the environment effect the expression of genes, but that epigenetic factors may be heritable (passed on to the next generation). Different cells have their own methylation pattern so that a unique set of proteins will be produced in order for that cell to perform its function. During cell division, the methylation pattern will be passed over to the daughter cell. In other words, the environment is affecting inheritance. Sperm and eggs develop from cells with epigenetic tags. When a sperm and egg cell meet to form a zygote, the epigenome (the sum of all the epigenetic tags) are removed through a process called “reprogramming”. About 1% of the epigenome is not erased and is passed on to the next generation. This is called “imprinting”. Ex: A pregnant mother may develop gestational diabetes (temporary diabetes while she is pregnant). As a result, high levels of glucose in the fetus can trigger epigenetic changes to the fetus’ DNA giving the child an increased chance of developing diabetes itself. Data Base Question: Changes in Methylation Pattern with age in Identical Twins One study compared the methylation patterns of 3-year olds identical twins with 50 year old identical twins Methylation patterns were dyed red on one chromosome for own twin and dyed grene for the other twin on the same chromsomes. The result would be a yellow colour if the patterns were the same. Differences in patterns on the two chromosomes result in green and red patches. This was done for 4 pairs of chromsomes. 1. Explain the reason for the yellow colouration if the methylation pattern is the same in the 2 twins. 2. Identify the chromosome with the least changes as the twins age. 3. Identify the chromosomes with the most changes as the twins age. 4. Explain how these differences could arise 5. Predict with a reason whether identical twins will become more or less similar to each other in their characteristics as they grow older.