slides for SMART-FHIR_rr_ga
advertisement

SMART/FHIR Genomic
Resources
An overview...
For latest see
http://smartgenomics.wikispaces.com/Clinical+Genomics+Presentation
Change Log
•
•
•
•
•
Made a few changes to Sequence resource
Added data support for alignment data (e.g. SAM or
BAM file)
VCFMeta, VCFVariant, GVFMeta, GVFVariant,
AlignmentMeta, AlignmentRead are exchanged using
REST path /binary, recognized as a Binary resource
SequencingLab replaced with an extension to
Procedure resource
GeneticAnalysis replaced with an extension to
Observation resource
List of Genomic Resources
Genetic Analysis
Implemented as an extension to Observation resource
Summary of genetic test
Documentation of phenotype-genotype association
Clinical decision support
•
•
•
•
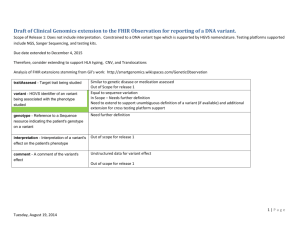
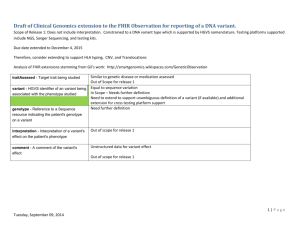
Extension to Observation resource
Added fields below to establish genotype-phenotype
association
● phenotype : CodeableConcept // 0..1 Target phenotype in the observation;
reason why the observation is performed
● variantObservation : {
identifier : string // 0..1 HGVS nomenclature of the variant
genotype : Resource(Sequence) // 1..1 reference to genotype
interpretation : CodeableConcept // 0..1 interpretation of genotype
comment : string // 0..1 comment...
} // 0..*
List of Genomic Resources
Raw data encapsulation (e.g. VCF Variant)
•
•
•
Encapsulates a row of raw genetic data (VCF, GVF,
SAM, BAM)
With reference to original file
Change note: We now support encapsulation of
alignment files SAM/BAM
List of Genomic Resources
Abstract representation - Sequence
•
•
•
An abstract of VCFVariant/GVFVariant
Enables developer to view genotypes without being
constrained by file formats
References raw data (e.g. reference to VCFVariant)
List of Genomic Resources
Sequencing Lab
•
•
•
“Folders” containing files of genetic data
Facilitates collaboration in research (bulks of data can
be shared via the API between various labs)
Implemented as an extension to Procedure resource
Extension to Procedure resource
Added fields below to document sequencing labs
● species : CodeableConcept // 1..1 species of the sample used in the
sequencing lab
● sample : {
type: code // 1..1 type of the sample used in the sequencing lab
source : CodeableConcept // 0..1 specific sample used
} // 1..1
● assembly : code // 0..1 assembly used in for alignment for analysis
● file : attachment // 0..* File generated from the lab for analysis
Use Case – Clinical Decision Support
•
•
•
•
Developer A has access to database documenting
genotype-phenotype association
Query Sequence resource with region of interest
Map the result against database and find out potential
risk factor of patient
Profile discovery of such risk factors with
GeneticObservation
Use Case – File Analysis
•
•
•
•
Developer B found some VCF files attached in
/procedure/123 as result generated from sequencing
lab
User can query for regions that he/she is interested in
using the API
VCFVariant – variant (genotypic) information
VCFMeta – legends that help user understand some
of the user-defined data within the variant info
Sequence
VCF Meta
VCF Variant
GVF Meta
GVF Variant
Alignment Meta
Alignment Read