What is raw data?
advertisement
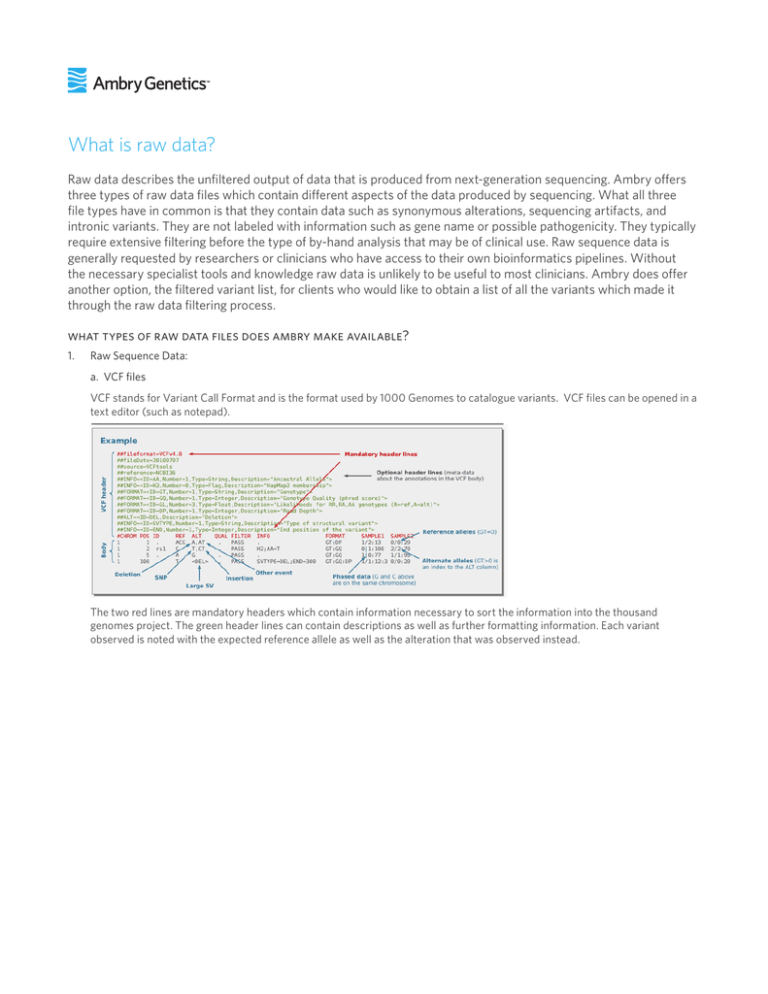
What is raw data? Raw data describes the unfiltered output of data that is produced from next-generation sequencing. Ambry offers three types of raw data files which contain different aspects of the data produced by sequencing. What all three file types have in common is that they contain data such as synonymous alterations, sequencing artifacts, and intronic variants. They are not labeled with information such as gene name or possible pathogenicity. They typically require extensive filtering before the type of by-hand analysis that may be of clinical use. Raw sequence data is generally requested by researchers or clinicians who have access to their own bioinformatics pipelines. Without the necessary specialist tools and knowledge raw data is unlikely to be useful to most clinicians. Ambry does offer another option, the filtered variant list, for clients who would like to obtain a list of all the variants which made it through the raw data filtering process. what types of raw data files does ambry make available? 1. Raw Sequence Data: a. VCF files VCF stands for Variant Call Format and is the format used by 1000 Genomes to catalogue variants. VCF files can be opened in a text editor (such as notepad). The two red lines are mandatory headers which contain information necessary to sort the information into the thousand genomes project. The green header lines can contain descriptions as well as further formatting information. Each variant observed is noted with the expected reference allele as well as the alteration that was observed instead. b. BAM files BAM files are a compressed version of SAM files. SAM stands for Sequence Alignment Map and is a format that contains the full sequence of each read that was successfully mapped. These files are very large and in order to be compressed these sequences are compressed into a binary version, the BAM files. Due to their compression, BAM files are unable to be read using text editors and require a special viewer such as the Integrative Genomics Viewer Overlapping segments are mapped and nucleotides which do not match up to the other overlapping segments are highlighted in red. Unlike the VCF format all nucleotides that are sequenced are represented here, not just variants. c. FASTQ files FASTQ files were developed to bundle a FASTA sequence along with quality data (FASTA + Q-Score= FASTQ). They are written in plain text and can be opened with a text editor like the VCF files. Each FASTQ file typically contains four lines per sequence. Like a FASTA file, Line 1 begins with an @ and contains a sequence identifier. It may also contain descriptive text. Line two contains the raw sequence letters. Line three begins with a + and may but does not necessarily contain the information from Line 1 repeated. Line 4 is composed of various ASCII symbols which represent varying qualities of sequence calls. 2. Filtered Variant List The filtered variant list is a combined list of variants remaining after bioinformatics filtering as well as manual filtering of identified sequencing artifacts, provided in excel format. These variants have not undergone interpretation and/or may represent sequencing artifacts as many have not been confirmed by a second laboratory method. This list of variants may include secondary findings. 50339.3083_v1 15 Argonaut, Aliso Viejo, CA 92656 Toll Free 866 262 7943 Fax 949 900 5501 ambrygen.com









