NNIN_highlights_examples_2
advertisement
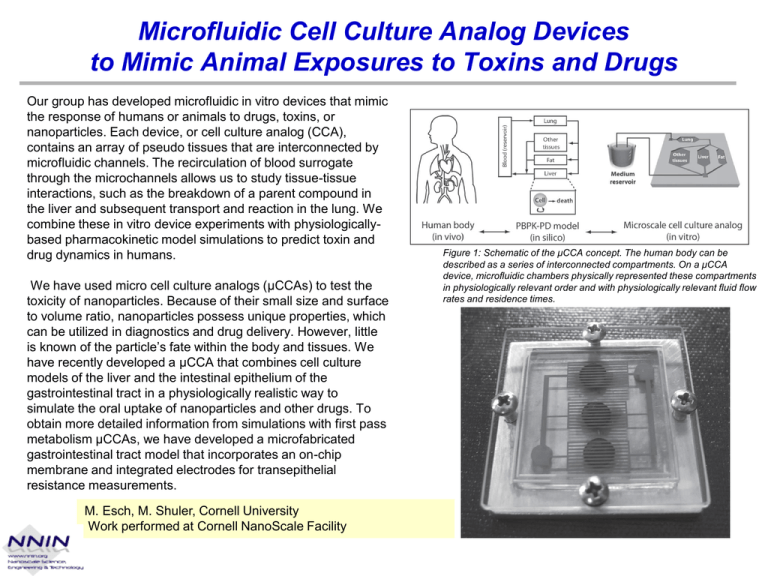
Microfluidic Cell Culture Analog Devices to Mimic Animal Exposures to Toxins and Drugs Our group has developed microfluidic in vitro devices that mimic the response of humans or animals to drugs, toxins, or nanoparticles. Each device, or cell culture analog (CCA), contains an array of pseudo tissues that are interconnected by microfluidic channels. The recirculation of blood surrogate through the microchannels allows us to study tissue-tissue interactions, such as the breakdown of a parent compound in the liver and subsequent transport and reaction in the lung. We combine these in vitro device experiments with physiologicallybased pharmacokinetic model simulations to predict toxin and drug dynamics in humans. We have used micro cell culture analogs (μCCAs) to test the toxicity of nanoparticles. Because of their small size and surface to volume ratio, nanoparticles possess unique properties, which can be utilized in diagnostics and drug delivery. However, little is known of the particle’s fate within the body and tissues. We have recently developed a μCCA that combines cell culture models of the liver and the intestinal epithelium of the gastrointestinal tract in a physiologically realistic way to simulate the oral uptake of nanoparticles and other drugs. To obtain more detailed information from simulations with first pass metabolism μCCAs, we have developed a microfabricated gastrointestinal tract model that incorporates an on-chip membrane and integrated electrodes for transepithelial resistance measurements. M. Esch, M. Shuler, Cornell University Work performed at Cornell NanoScale Facility Figure 1: Schematic of the μCCA concept. The human body can be described as a series of interconnected compartments. On a μCCA device, microfluidic chambers physically represented these compartments in physiologically relevant order and with physiologically relevant fluid flow rates and residence times. The Evolutionary Dynamics of Drug Resistance by Drug-induced Stress Gradients Emergence of resistance to antibiotics by bacteria is a growing problem, yet not well understood. In a microfluidic device designed to mimic naturally occurring bacterial niches. Evolution proceeds most rapidly with just the right combination of a large number of mutants and rapid fixation of the mutants (Goldilocks point). Using a two-dimensional micro-ecology it is possible to fix resistance to the powerful antibiotic ciprofloxacin (Cipro) in wildtype E. coli in 10 hours through a combination of extremely high population gradients, which generate rapid fixation, convolved with the “just right” level of antibiotic which generates a large number of mutants and the motility of the organism. The design of the micro-ecology is unique in that it provides two overlapping gradients, one an emergent and self generated bacterial population gradient due to food restriction and the other a mutagenic antibiotic gradient. Further, it exploits the motility of the bacteria moving across these gradients, to drive the rate of resistance to Cipro to extraordinarily high rates. Whole genome sequencing of the resistant organisms revealed four functional single nucleotide polymorphisms attained fixation. Rapid emergence of antibiotic resistance in the heterogeneous conditions prevailing in the mammalian body may also apply to the emergence of drug resistance during cancer chemotherapy. G. Lambert, R. H. Austin, Princeton University Work performed at Cornell NanoScale Facility (A) An overview of the entire micro-ecology, showing the flow of the nutrient streams and the nutrient+Cipro containing streams. The nutrient stream is x1 LB broth, while the nutrient+Cipro stream is x1 LB broth + 10 μg/mL Cipro. (B) Image of the expected Cipro concentration using the dye fluorescein as a marker. The asymmetry of the pattern at the Goldilocks points is due to the direction of the flow. (C) Scanning electron microscope (SEM) image of the area of the array in (A) outlined by the box. Each hexagon is etched down 10 microns, the interconnecting channels are 10 microns deep and 10 microns wide. (D) SEM image of the nanoslits at the micro-ecology periphery. The nanoslits are etched down 100 nm and are 6 microns wide and 10 microns long. Proc. of SPIE Vol. 7929 (2011)