slides - Smith Lab
advertisement
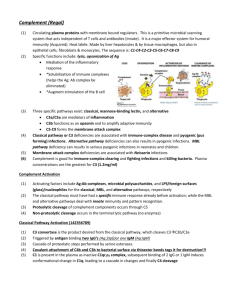
Immunology of Age Related Macular Degeneration Kyle C. McKenna, Ph. D. Associate Professor of Biology, Franciscan University of Steubenville Associate Professor of Ophthalmology, University of Pittsburgh mckennakc@upmc.edu 412-802-8437 Age Related Macular Degeneration Leading cause of blindness in individuals over the age of 60 Due to atophy of the macula area of the retina where central vision is focused. Retinal Architecture Bruch’s membrane Photoreceptors RPE Choroid Sclera Macular Degeneration Types • Dry Form – Loss of RPE and overlying retina resulting in “geographic atrophy” • Wet Form – Neovascularization of macula, inflammation, retinal scarring, associated with severe vision loss AMD Pathogenesis Normal Aging causes Thickening of Bruch’s Membrane Toxic products of Phototransdution Accumulate in RPE RPE cell death Dry AMD AMD Pathogenesis Thickening of Bruch’s Membrane Toxic products of Phototransdution Accumulate in RPE Choroidal neovascularization, Retinal edema Scar formation RPE cell death Wet AMD AMD Risk Factors •Age. In the United States, macular degeneration is the leading cause of blindness in people age 60 and older. •Cigarette smoking. Exposure to cigarette smoke doubles the risk of macular degeneration. •Low levels of nutrients. This includes low blood levels of minerals, such as zinc, and of antioxidant vitamins, such as A, C and E. Antioxidants may protect cells from oxygen damage (oxidation), which may partially be responsible for the effects of aging and for the development of certain diseases such as macular degeneration. •Family History of AMD. SNP associations in AMD • SNP Y402H in Complement Factor H associated with • increased risk • SNP LOC 387715 which localizes to a mitochondrial protein is associated with increased risk Het. At either Y402H or Loc 387715: 2.8 fold risk Het At both Y402H or Loc 387715: 5.8 fold risk Hom At either locus: Y402H: 7.1 risk, Loc 387: 8.1 Hom at both loci: 57-fold risk What is the Complement? • Collection of heat-labile soluble proteins constitutively produced primarily by the liver that are found in blood, lymph and extracellular fluids – – – – C1 (C1q:C1r2:C1s2), C4(C4b, C4a), C2(C2a, C2b), C3 (C3a,C3b) Collectins (MBL) Ficolins Factor D, Factor B, Properdin C5 (C5a, C5b), C6, C7, C8, C9) • Many complement proteins are proteases that are synthesized as inactive pro-enzymes (zymogens) – C1(C1q:C1r2:C1s2) – Collectins or Ficolins (MASP1, MASP2) What is the function of Complement? • Opsonization to promote phagocytosis • Stimulation of inflammation • Clearance of Immune Complexes, apoptotic cells, cellular debris • Lysis of bacteria, viruses, and cells that are damaged or infected. Opsonization & Phagocytosis • Complement activation deposits C3b on the surface of bacteria, viruses, or cells that are infected or damaged • C3b receptors are present on phagocytes – CR1 (CD35): C3b macrophages, neutrophils, RBC – CR2 (CD21): C3dg macrophages and B cells – CR3 (CD11b): iC3b macrophages and neutrophils – CR4 (CD11c) : iC3b dendritic cells CR1 Binds C3b and Augments Phagocytosis (Not Janeway) Phagocytosis may require additional activation via C5aR : C5 C3aR : C3 Opsonization & Phagocytosis • Phagocytosis is enhanced by antibodies binding to Fc receptors on the surface of phagocytes • Hence, the antibody response (adaptive) is complemented by the innate response (complement) Opsonization B Menu F Inflammation • Certain complement proteins generated during the complement cascade are inflammatory (anaphylotoxins) – C3a – C4a – C5a Potency C5a>C3a>C4a Inflammation Increase vascular permeability Promote Chemotaxis of Immune Cells (neutrophils) Promote Mast Cell Degranulation Increased phagocytosis Removal of Immune Complexes • Complement also helps clear immune complexes (Antigen/Antibody conjugates) • Immune complexes can cause many problems, esp. in autoimmune diseases • Immune complexes bind C3b • Recognized by CR1, esp. on RBC’s • Phagocytosed in spleen and liver Functions of the Complement System © Removal of Immune Complexes via C3b and the CR1 receptor Removal of Apoptotic Cells • Phosphocholine, present in bacterial phospholipids is recognized by C-reactive protein • C-reactive protein activates complement • Apoptotic cells express phosphocholine on the cell surface which activates complement for subsequent removal (d) B Menu F MAC lysis • The MAC pore size is 70-100Å • Allows ions and small molecules to diffuse out • Disrupts osmotic stability and lyses bacteria, virus, or cells that are infected or damaged MAC Pores Summary of Complement Functions In addition to providing protection from pathogens, complement plays an Important role in normal physiology by removing apoptotic and damaged cells and cell debris How is Complement Cascade Initiated? • By Direct Binding of Pathogen Surfaces (Mannose/Lectin Pathway) • By Indirectly Binding of Pathogen Surfaces (Classical Pathway) via engagement of antibodies or C-reactive protein • Spontaneously (Alternative Pathway) INITIATION ACTIVATION AMPLIFICATION EFFECTORS INITIATION: Classical Pathway • C1 complex can bind: C1 complex – Antibodies complexed with antigens • Natural • Adaptive – C-reactive protein • Acute phase protein that binds phophocholine in bacterial polysaccharides and apoptotic cells – Microbe surfaces • Lipoteichoic acid (Gm+) • Some bacterial proteins INITIATION: Classical Pathway Not Janeway -The initial steps in classical pathway initiation are very similar when C1 binds C-reactive protein (or a pathogen surface). INITIATION: Classical Pathway -NOTE: Cleavage of complement proteins yields “a” and “b” products. -The “b” product is ALMOST ALWAYS the larger (big) one and binds to the surface INITIATION: Classical Pathway FORMATION OF C3 CONVERTASE The exception to the “b” rule C4b2a is a C3 convertase C3b deposition on cell surface INITIATION: Lectin Pathway - Present in low concentrations in plasma; production by liver increased during acute phase response - Two- to six-headed that forms complex with two protease zymogens - MASP-2 closely related to C1r and C1s INITIATION: Lectin Pathway Ficolins have a fibrinogen-like domain That binds acetylated sugars but not Mannose n-linked glycoproteins are terminated with sialic acid residues. Complement activation highly similar to classical pathway -Upon binding a pathogen surface, a conformational change in MASP-2 occurs resulting in cleavage of C4 and C2, formation of C3 convertase (C4bC2a) -C3b is deposited on the cell surface INITIATION: Alternative Pathway -In plasma, circulating C3 spontaneously undergoes hydrolysis to form C3(H2O) which binds factor B -Factor B on C3(H2O) is cleaved by Factor D to form C3(H2O)Bb, a C3 convertase -C3b is deposited on the cell surface INITIATION: Alternative Pathway FORMATION OF C3 CONVERTASE -Factor B binds C3b on cell surface which is subsequently cleaved by Factor D to form C3bBb, a C3 convertase -C3 convertase is stabilized by Factor P (properdin) (-properdin: t1/2 = 5 min; + properdin; t1/2 = 30 min) Properdin (Factor P) is made by neutrophils AMPLIFICATION Classical & Lectin C3 convertase Alternative C3 convertase -One molecule of C3 convertase can cleave up to 1000 molecules of C3 into C3b -Many C3b molecules deposited on cell surface (2 x 106 C3b molecules deposited in <5 min) EFFECTOR (Assembly of Membrane Attack Complex (MAC)) Formation of C5 convertase Cleavage of C5 convertase Classical/Lectin Alternative First step in MAC assembly EFFECTOR (Assembly of Membrane Attack Complex (MAC)) All complement pathways converge to this process Hydrophobic Domain Subsequent steps in MAC assembly C5b is labile and will be inactivated w/i two minutes unless stabilized by C6 EFFECTOR What happens to the complement fragments not deposited on the surface? ANAPHYLATOXINS -C3a, C4a and C5a are anaphylatoxins that induce local inflammatory responses Summary Complement Regulation Also, C4 (Not Janeway) -C3b, C4b rapidly inactivated by water unless allowed to immediately bind to protein or carbohydrate on cell surface Complement Regulation • C1 inhibitor (C1INH). – Dissociates C1r and C1s from C1 thereby removing the enzymatic activity necessary to cleave C4 and continue the complement cascade Complement Regulation Complement Regulatory Proteins Found in Plasma C4BP displaces C2a from the C4b2a complex. C4b bound by C4BP is cleaved by a soluble Factor I to inactive forms C4d and C4c C4d C4c -Factor I cleaves C4b and C3b but requires cofactors to yield inactive proteins: -C4 binding protein (C4BP) aids Factor I in cleaving C4b Complement Regulation Complement Regulatory Proteins Found in Plasma (Not Janeway) -Factor H also acts as a cofactor to Factor I to cleave C3b to yield inactive protein Complement Regulation Complement Regulatory Proteins Embedded in Cell Membranes (Not Janeway) -Decay-accelerating factor (DAF) and Membrane Cofactor Protein (MCP) prevent formation of convertases by displacing C3b -MCP further acts as a cofactor to Factor I to yield inactive C3b -Other: CR1 also prevents formation of C3 convertase by displacing C2a and/or Bb and acts as cofactor to Factor I to cleave C4b and/or C3b Complement Regulation Complement Regulatory Proteins Embedded in Cell Membranes -CD59 (protectin) prevents complete assembly of MAC Unregulated Complement in AMD B-amyloid in Drusen inhibits Factor I cleavage of C3b Wang J. et al. 2008. J. Immunol. 181:712 mRNA expression of Factor H by RPE is reduces by Oxidative stress (smoking). Wu Z. et al. 2007. J. Biol. Chem. 282:22414 Phagocytosis of oxidized photoreceptor cells by RPE Inhibits Factor H production by RPE Chen M. et al. 2007. Exp. Eye Res. 84:635 Y402H Polymorphism Effects • Y402H Polymorphism in Factor H decreases the affinity of Factor H for CRP – Prossner B. E. et al. 2007. J. Exp. Med. 204:2277 • Y402H Polymorphism in Factor H decreases the affinity of Factor H for the oxidized lipoprotein malondialdehyde (MDA) – Weismann D et al. 2011 Nature 478:76-81 CFH mutations prevent binding of CFH to cell surface thereby causing unregulated Complement activation on the cell surface leading to retinal inflammation Inflammasome Inflammasome activation is associated with geographic atrophy in AMD patients Tarallo V et al. 2012 Cell 149:847-859 Photoreceptors Apoptic Cell RPE C4b C4 C4b2a C2 C3 C3b C3bBb C3b B C4a C2b C4b2aC3b D C5 C5b C6 – C9 C5a C3a C3b2Ba Ba Oxidized Phospholipids Drusen BM Choroid Mac Mac Mac VEGF Photoreceptors Binds to CRP RPE C4b2a C2 C4b C3 C3b C3bBb C3b B C4 C4a C2b C4b2aC3b D C5 C5b C6 – C9 C5a C3a C3b2Ba Ba Inhibits C3 convertase formation Masks oxidized phospholipids FACTOR H Drusen BM Choroid Mac Mac Mac Retinal Damage • Induces anti-retinal antibodies along with antibodies to carboxypyrole adducted phospholipids