Genetic predictors of lung cancer
risk and progression
Some results and new proposals
Christopher Amos, Ph.D.
Olga Gorlova, Ph.D.
Ivan Gorlov, Ph.D.
Konstantin Dragnev
Scott Gerber, Ph.D.
James Rigas, M.D.
David Christiani, M.D, Sc.D.
Genetic Associations and Mechanisms in
Oncology (GAME-ON):
Transdisciplinary
Studies of Genetic
Variation in
Follow-up of
Ovarian
Cancer
Colorectal
Cancer
Genetic Association
and Interaction
Studies (FOCI)
Thomas Sellers
(CORECT)
Stephen Gruber
Elucidating Loci
Involved in
Prostate
Cancer
Susceptibility
(ELLIPSE)
Brian Henderson
PHASE 1
DISCOVERY
Transdisciplinary
Research in Cancer of
the
Lung
(TRICL)
Chris Amos
PHASE 2
FUNCTIONAL
ANALYSIS
Discovery, Biology, and
Risk of Inherited
Variants in
Breast
Cancer
(DRIVE)
David Hunter
PHASE 3
RISK ASSESMENT
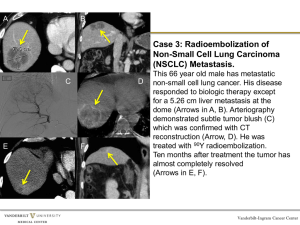
Manhattan plot of all lung cancers
from 1000 Genomes Imputation
CHRNA5
hTERT
BRCA2
TP63
hMSH5
CHEK2
Associations of common mutations
in BRCA2 with cancer in Iceland
Comparison of AD and SQ LC
Adenocarcinoma
Squamous Carcinoma
CHRNA5
(C) SQ
hTERT
TP63
CHRNA5
BRCA2 CHEK2
hMSH5
CLPTM1L CDKN2 RAD52
Squamous Lung Cancer
BRCA2
BRCA2
CHEK2
CHEK2
GWAS-translation
• Knockdown studies of CLPTM1L and TERT show loss
of CLPTM1L expression is necessary for lung cancer
development in a kRAS knockout mouse
• Comprehensive promoter methylation studies of risk
loci implicate epigenetic deregulation of most SNPassociated lung cancer loci including CHRNA3,
CHRNB4 and TERT in lung cancer susceptibility
• Genotype-methylation associations in lung tumor tissue for TERT and CHRNB4
• CHRNB4 promoter hypomethylation and CHRNA3 + TERT promoter
hypermethylation as well as methylation-expression correlations in tumor tissue
• CHRNB4 knockdown leads to reduced proliferation and propensity to form
colonies
Other Genetic Analysis Projects
Custom Affymetrix Array
• 9 studies concentrating on
cohorts
– 7,500 lung cancer cases
– 7,500 controls
Custom Array with 300,000
exome array markers
100,000 custom markers
including markers derived from
sequencing studies and
pharmacogenetic variants
Exome plus targeted regions
sequencing
• Sequencing of 1000 lung
ca. cases and 1000 contols
• Funded through a separate
application to CIDR
• Includes samples from the
Custom Affymetrix Array
Study to inform imputations
• Selecting early onset cases,
family history positive,
cases with tumor samples
and rare variant carriers
GAME-ON OncoArray
Common Content – 40K
Fine-mapping of common cancer susceptibility loci (TERT, 8q24 (proximal and
distal to MYC), HNF1B, TET2, RAD51B, 11q13, MERIT40, MDM4)
Ancestry Informative Markers
Cross-Site meta analysis
Pharmacogenetic components
eQTL (Height, Weight, BMI, WHR, Menarche, Menopause etc)
Other cancers published GWAS variants
Chromosome X and mitochondrial DNA variants
GWAS Backbone
260K
Illumina Core
OncoChip
600K
beadtypes
Cancer Specific
Variants
Lung
Colon
Breast
Prostate
Ovarian
(proportional allocation)
Proposed Research Studies
• Shared decision making and tumor analysis
– Proposed application of 100 lung cancer
cases with hotspot mutation versus exome
sequencing
• Collaboration between Karmanos Cancer
Institute and Dartmouth
• Reviewers liked Dartmouth component but
not Karmanos – lack of electronic medical
record at KCI, insufficient process details
Predicting Risk for Recurrence
• Proposed collaboration to Lungevity
Foundation
• Uses snap frozen samples from Harvard to
perform integrated analysis – genomic
mutations and proteomic alternations
• 200 cases selected for recurrence or
nonrecurrence
• Could be extended in R01 to larger sample size
• Extend to other lung cancer phenotypes
U01 Grant On Integration of SNP Data in
Lung Cancer Screening
• In collaboration with Dr. Kimmel from Rice we are working on the
proposal to integrate GWAS-detected risk and outcome SNP into
lung cancer screening model.
• As the first step we will estimate effects of SNPs on tumor
growth and metastasizing rate. We will use NLST and TCGA data.
• We will then incorporate SNPs into the model of natural history
of lung cancer with the screening module superimposed onto it.
• SNPs in the model will be incorporated based on their frequency
and estimated effect size on tumor growth and metastasizing
rates.
• The goal is to estimate if targeted genotyping of the risk
associated SNPs will improve screening efficacy.
P01 Integrative Analysis of Lung
Cancer Risk
Project 1
Smoking
Genetic Predictors
Dependence
Project 4:
Application of Risk
Models to
Screening
Populations
Biostatistics
and QC Core
Project 2: Genomic
and Epigenetic
Predictors of Risk
Project 3:
Intermediate
predictors of risk:
miRNA,
metabolomiic and
‘nutritional’
exposures
Genomics and Genetics
Genetic Mapping of DNA Methylation in EAGLE Lung
Conducted methylation quantitative trait loci (QTL) analysis of
EAGLE normal lung tissues in 210 samples, with 450K CpG
probes, replicated in TCGA lung tissue
(Additive model between each SNP and normalized methylation trait pair, adjusting
for sex, age, plate, population stratification and methylation-based PCA scores)
34,304 cis-meQTL
(mapping to 9,963 genes)
cis region=500kb
585 trans-meQTLs
trans region>500kb or
different chromosomes
Most meQTLs are not in gene promoters or CpG islands
Shi et al., Nature Communications (In press)
CHRNA5
CLPM1L
TERT
15q25
6p21
5p15
12p13
9p21
CHRNA3
RAD52
CDKN2A
MSH5
Inherited genetic variation may affect lung carcinogenesis by
cis-meQTL
in lung cancer GWAS loci
regulating the
human methylome