I. Comparing genome sequences
advertisement

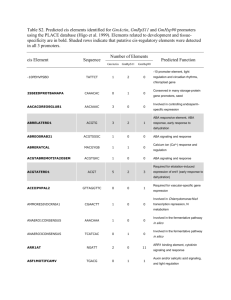
Comparative Genomics II: Functional comparisons Caterino and Hayes, 2007 Overview I. Comparing genome sequences • Concepts and terminology • Methods - Whole-genome alignments - Quantifying evolutionary conservation (PhastCons, PhyloP, GERP) - Identifying conserved elements • Utility and limitations of conservation • Available datasets at UCSC II. Comparative analyses of function • Evolutionary dynamics of gene regulation • Case studies • Insights into regulatory variation within and across species Functional variation within and among species Human Chim p Rhes us Mous e Regulatory variation contributes to human phenotypic variation Modularity of developmental gene expression overall Limb TFs gene A limb gene A forebrain Brain TFs Neural TFs gene A neural tube Regulatory changes introduce variance without disrupting protein Regulatory mutations affecting pleiotropic genes cause discrete developmental changes Lettice et al. Hum Mol Genet 12:1725 (2003) Sagai et al. Development 132:797 (2005) Patterns of selection on gene expression and regulation Neutral Constrained Romero et al., Nat Rev Genet. 13:505 (2012) Directional Comparative approaches to identify conserved and variant regulatory functions Regulatory conservation Regulatory rewiring Visel and Pennacchio, Nat Genet 42:557 Genetic drivers of gene regulatory variation Furey and Sethupathy, Science Comparative analysis of ChIP-seq datasets • H3K4me2 • H3K27ac Human Mouse • H3K4me2 • H3K27ac Compare TF binding, histone modifications, DNase hypersensitivity in equivalent tissues Requires a statistical framework to reliably quantify changes in ChIP-seq signals Issues in comparative functional genomics •Input data are noisy: ChIP-seq, RNA-seq data are signal based, sub to considerable experimental variation •Using comparable biological states within and across species (e.g., human liver vs. mouse liver) = variation across tissues? •How do epigenetic states and gene expression diverge among individuals and across species (Neutral? Constrained?) •Can we identify variants or substitutions that drive regulatory changes? Science 328: 232 (2010) •Targets: RNA Polymerase II NFkB •10 human lymphoblastoid cell lines 3 major population groups: European, East Asian, Nigerian 9 females, 1 male 9 analyzed by HapMap and 1000 Genomes NFkB PolII Fraction of regions bound Pairwise difference in binding Variation in TF binding is common # individuals Science 342: 747 (2013) •Targets: RNA Polymerase II H3K4me1, H3K4me3, H3K27ac, H3K27me3 DNase hypersensitivity •10 human lymphoblastoid cell lines 1 population group (Nigerian) All analyzed by HapMap and 1000 Genomes Measuring allelic imbalance in histone modification profiles ChIP-seq reads G allele Allelic imbalance T allele Need to map reads reliably to individual alleles Cis-quantitative trait loci ~1200 identified Science 328: 1036 (2010) •Targets: CCAAT/enhancer binding protein a (CEBPA) Hepatocyte nuclear factor 4a (HNF4A) Essential for normal liver development and function •Tissue: Adult liver from 4 mammal species plus chicken Lineage-specific gain and loss of CEBPA binding in liver Lineage-specific: 0 bp overlap in multiple species alignment Widespread variation in CEBPA binding in mammals Widespread variation in CEBPA binding in mammals Cell 154: 530 (2013) Single TF binding events may not indicate regulatory function • Many TFs are present at high concentration in the nucleus • TF motifs are abundant in the genome Enhancer-associated histone modification • Single TF binding events may be incidental Combinatorial TF binding events are more conserved Many TF binding changes do not have obvious genetic causes In mammalian liver: Many TF binding changes do not have obvious genetic causes In mouse liver: Cell 154: 185 (2013) Human Rhesus Mouse Bud stage; digit specification Digit separation Identifying human-lineage changes in promoter and enhancer function • Compare H3K27ac signal at orthologous sit • ‘Stable marking’: 1.5-fold or less change in H3K27ac among human, rhesus and mou • Human gain: require significant, reproducibl gain in human versus all 12 datasets in rhesus and mouse Mapping active promoters and enhancers in human limb ENCODE cell lines H3K27ac Gains in promoter and enhancer activity • Bone morphogenesis • Chondrogenesis • Digit malformations in mouse Human-specific H3K27ac marking correlates with changes in enhancer function Epigenetic signatures reflect tissue identity and species relationships H3K27ac signal in human and mouse H3K27ac in human, rhesus, mouse Primate Mouse Nature 478: 343 (2011) • • • • • • • • • • Human Chimpanzee Bonobo Gorilla Orangutan Macaque Mouse Opossum Platypus Chicken • • • • Custom gene models based on Ensembl + RNA-s 5,636 1:1 orthologs in amniotes 13,277 1:1 orthologs in primates Only constitutive exons Global patterns of gene expression differences Gene expression recapitulates species phylogenies Gene expression divergence rates are tissue-specific testis liver brain Gene expression divergence increases with evolutionary time Conservation of core organ functions restricts divergence Summary •Comparative functional genomics identifies regulatory differences within and among species •TF binding is variable within species and highly variable among species •Epigenetic comparisons provide more insight into biologically relevant regulatory diversity and divergence •Gene regulation and expression diverges with increasing phylogenetic distance – they mirror neutral expectation