ch5_SP13x
advertisement
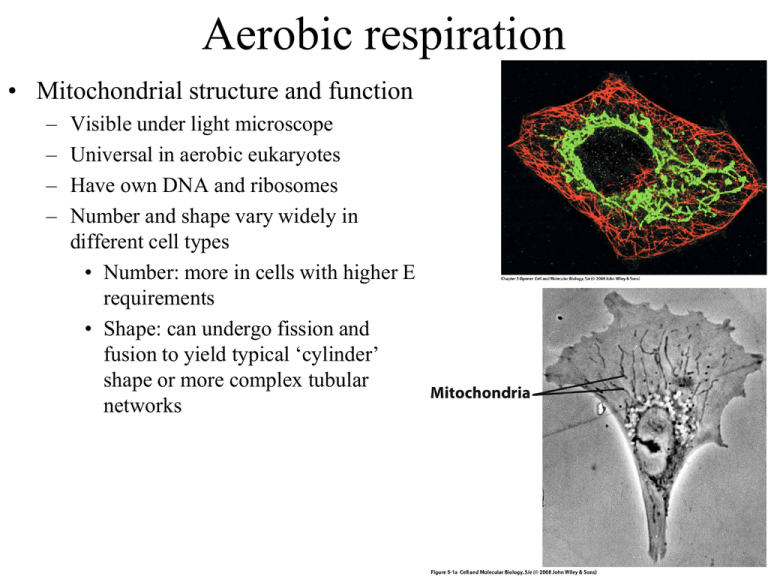
Aerobic respiration • Mitochondrial structure and function – – – – Visible under light microscope Universal in aerobic eukaryotes Have own DNA and ribosomes Number and shape vary widely in different cell types • Number: more in cells with higher E requirements • Shape: can undergo fission and fusion to yield typical ‘cylinder’ shape or more complex tubular networks Aerobic respiration • Mitochondrial structure and function – Membranes • Outer: permeable to many things – Porins, large central pore • Inner: highly impermeable – Channels for pyruvate, ATP, etc Aerobic respiration • Mitochondrial structure and function – Membranes • Outer: permeable to many things – Porins, large central pore • Inner: highly impermeable – Channels for pyruvate, ATP, etc • Cristae – Complex invaginations of the inner membrane – Functionally distinct – Joined to inner membrane via narrow channels Aerobic respiration • Mitochondrial structure and function – Intermembrane space • Between inner and outer membranes • Also within the cristae • Acidified ( high [H+] ) by action of the Electron Transport Chain (ETC) – H+ are pumped from matrix into this compartment – ATP synthase lets them back into the matrix Aerobic respiration • Mitochondrial structure and function – Matrix • Compartment within the inner membrane • Very high protein concentration ~500mg/ml • Contains: – ribosomes and DNA – Enzymes of TCA cycle, enzymes for fatty acid degradation NADH enters the mitochondria by one of two mechanisms: 1. aspartate-malate shuttle NADH --> NADH 2. glycerol phosphate shuttle NADH --> FADH2 • Pyruvate to TCA Oxidation-reduction potentials • Reducing agents give up electron share – The lower the affinity for electrons, the stronger the reducing agent • NADH is strong, H2O is weak • Oxidizing agents receive electron share – The higher the affinity for electrons, the stronger the oxidizing agent • O2 is strong, NAD+ is weak • Couples – NAD+ - NADH couple – O2 - H2O couple (weak oxidizer, strong reducer) (strong oxidizer, weak reducer) strong oxidizing strong reducing NADH is a stronger reducing agent than FADH2 G = -nFE NADH --> H2O G0’= -52kcal/mol 7ATP(max), ~3ATP(real) FADH2 --> H2O G0’= -36kcal/mol 5ATP(max), ~2ATP(real) The Tricarboxylic Acid (TCA) cycle (Kreb’s cycle) 2Pyruvate + 8NAD+ + 2FAD + 2GDP + 2Pi --> 6CO2 + 8NADH + 2FADH2 + 2GTP Adding in products of glycolysis, 2NADH + 2ATP Total yield for both: 10NADH + 2FADH2 + 4ATP = 38 ATP How NADH from cytoplasm are counted changes the theoretical yield Formation of a tricarboxylic acid from pyruvate • In two steps: – A dehydrogenase step 3C + NAD 2C + CO2 +NADH – Yields Acetyl group bonded to CoenzymeA (CoA) – A synthase step – 2C + 4C(OA) 6C (OA) The Tricarboxylic Acid (TCA) cycle (Kreb’s cycle) • 2C+4C(OA) 6C • 6C+NAD 5C+CO2 +NADH • 5C 4C+CO2 +NADH • 4C+GDP 4C+GTP • 4C+FAD 4C+FADH2 • 4C+NAD 4C(OA) +NADH Fatty acid catabolism • Enzymes localized to mitochondrial matrix – Fatty acids cross inner membrane and become linked to HS-CoA – Each turn of cycle generates FADH2 + NADH2 + Acetyl-CoA Amino acid catabolism • Enzymes in mitochondrial matrix – cross inner membrane via specific transporters – Enter TCA at various points General outline of oxidative phosphorylation Electron Transport Chain: e- carriers • Electron carriers – Flavoproteins (FMN) – Ubiquinone (Q or UQ) – Cytochromes (b, c1, c, a) – Cu atoms – Fe-S centers – Proton movement driven by complexes I, III, IV coupled to large E Electron carriers: Ubiquinone • Lipid soluble • Dissolved within inner mitochondrial membrane • Free radical intermediate Q • Free radical ‘escape’ from electron transport chain can damage proteins, lipids, RNA, and DNA in a cell UQ Electron Transport Chain • Complex I passes e- from NADH to Q and pumps 4H+ out of matrix • Complex II passes e- from FADH2 to Q • UQ shuttles e- to Complex III Electron Transport Chain • Complex III passes e- to Cytochrome c and pumps 4H+ out of matrix • Cytochrome c passes e- to Complex IV • Complex IV passes e- to O2 forming H2O and pumps 2H+ out 1 pH unit diff ATP synthesis: The ATP Synthase enzyme • F1 head/sphere (ATPase) catalyzes ADP + Pi <--> ATP • F0 base embedded in inner membrane (H+ pass through this) • F0 + F1 = ATP synthase – Connected via two additional proteins • Central rod-like gamma subunit • Peripheral complex (abd) holds F1 in a fixed position – Location • Bacteria = plasma mem • Mitochondria = inner mem • Chloroplast = thylakoid ATP matrix H+ Intermembrane space Binding Change mechanism of ATP Synthase • Each F1 active site progresses through three distinct conformations – Open (O) Loose (L) Tight (T) – Conformations differ in affinity for substrates and products • Central gamma () subunit rotates causing conformation changes Rotational catalysis by ATP synthase • Central gamma () subunit rotation caused by proton (H+) translocation drives the conformation changes 1 pH unit diff Rotational catalysis by ATP synthase • If true, should be able to run it backwards (ATP --> ADP + Pi) and watch gamma spin like a propeller blade Rotational catalysis by ATP synthase Other fxns of electrochemical gradient • E also used for: – Import of ADP + Pi (+H+) and export of ATP – Import of pyruvate (+H+) • Uncoupling sugar oxidation from ATP synthesis – Uncoupling proteins (UCP1-5) • UCP1/thermogenin, shuttles H+ back to matrix (endothermy) – Brown adipose tissue » Present in newborns (lost with age) and hibernating animals » Generates heat – 2,4-dinitrophenol (DNP) • Ionophore that can dissolve in inner membrane and shuttle H+ across – 1930’s stanford diet pill trials: overdose causes a fatal fever