Epigenetic Studies in ALSPAC
advertisement

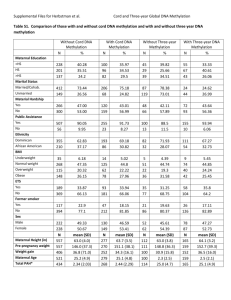
Epigenetic Studies in ALSPAC Caroline Relton CAiTE Symposium 12th January 2010 Objectives • Define DNA methylation variation due to DNA source and extraction method • Quantify changes in DNA methylation over time • Validate differential DNA methylation at selected target loci in serial DNA samples • Undertake pilot MeDIP-sequencing of the entire methylome • Quantify differential DNA methylation in pre- and post-menopausal women • Apply a Mendelian randomisation approach to strengthen evidence for a causal relationship between environmental exposures, DNA methylation and childhood outcomes • Develop bioinformatic and statistical approaches for handling DNA methylation data Define DNA methylation variation due to DNA source and extraction method Time Tube Extraction T1 Birth Heparin Phenol T2 43 months EDTA Phenol T3 61 months EDTA Phenol T4 7 years EDTA Salting out T5a 9 years CPD/ACD Guanidine hydrochloride T5b 9 years Cell ine Guanidine hydrochloride T0 Pregnancy EDTA or heparin Phenol T0+17 +17y follow-up EDTA Guanidine hydrochloride Avg Beta, Phenol Average beta (Me/unMe) # Buffy coat White cells Whole blood 114 probes more methylated in guanidine extracted DNA (> 1.5-fold) Avg Beta, Guanidine Quantify changes in DNA methylation over time • Sequenom EpiTyper • 6 amplicons • 12-21 CpG sites per amplicon • Birth and 7y DNA • N=90 • Correlation is much lower than that observed in adults at 2 time points • Additional samples are being analysed Highest intra-probe correlations Amplicon Spearman’s rho (B) 95% CI (B) Spearman p FTO_16 0.341 0.099, 0.568 0.006 P16_38 0.292 0.075, 0.490 0.008 P16_4 0.265 0.058, 0.458 0.014 IGF2BP2_11 -0.242 -0.439, -0.04 0.024 IGF2BP2_29 0.237 0.003, 0.464 0.039 PPARg_29 0.215 0.020, 0.398 0.040 P16_25 0.211 0.014, 0.397 0.047 IGF2BP2_26 0.216 -0.017, 0.443 0.047 B = bootstrapped Validate differential DNA methylation at selected target loci BMI Fat mass SNP-dependent locus associated with insulin resistance Methylation at CpG vs rs231840 Lean mass CDKN1C MMP9 MPL CDKN1C EPHA1 HLA-DOB NID1 Methylation (%) 6.0 5.0 4.0 3.0 2.0 1.0 0.0 -1.0 -2.0 -3.0 CASP10 CDKN1C EPHA1 Change in outcome / 1% in methylation Methylation at birth and body composition in childhood Genotype (rs231840) Undertake pilot MeDIP-sequencing of the entire methylome • High vs normal BMI • Aged 0y, 7y and 15y • N=10 per group • MeDIP-seq pilot • Illumina 27K array Quantitative comparison of genome-wide DNA methylation mapping technologies Christoph Bock, Eleni M Tomazou, Arie B Brinkman, Fabian Müller, Femke Simmer, Hongcang Gu, Natalie Jäger, Andreas Gnirke, Hendrik G Stunnenberg & Alexander Meissner Nature Biotechnology : 28: 1106–1114 (2010) Quantify differential DNA methylation in pre- and post-menopausal women Pregnancy vs +17y Avg beta Post MP ▫ 1032 CpG sites differ +/- 5% Avg beta Pre MP Pre vs Post menopause ▫ 199 CpG sites differ +/- 5% Average methylation for 5 largest methylation shifts in each direction with SD error bars Apply a Mendelian randomisation approach CVD BMI CpG ADH1B Reverse causation CVD Confounded Alcohol CpG HNSCC CpG BMI CpG CVD On causal pathway Socio-economic position Nutritional status Smoking BMI CVD CpG Independent and both causal Alternative non-epigenetic pathway Develop bioinformatic and statistical approaches Defining where in the genome to look for differential methylation • In silico tools ▫ CGI Explorer ▫ Data mining tools ▫ Transcription factor binding sites • Gene expression data • Whole methylome analysis ▫ MeDIP-seq • Genome-wide site-specific analysis ▫ Illumina 450k array • Targeted approaches ▫ Illumina VeraCode Analysing DNA methylation data • Large data sets • Highly correlated • Non-normal distribution • Outlier effects • Temporal variation • Tissue specificity • Differences in DNA source and method used ▫ Illumina ▫ Sequenom ▫ Pyrosequencing • Relationship between genotype and epigenotype Grant submissions and future plans • Grants awarded ▫ ▫ ▫ WT/MRC Strategic Award (GDS) WT ALSPAC Mums (DAL) MRC Fellowship (LZ) • Grant submitted ▫ ▫ ▫ ▫ NIH Conduct problem trajectories (EB) NIH Obesity and epigenetics (CR) MRC ALSPAC Mums (DAL) BBSRC BBR (GDS) • Grants in preparation ▫ ▫ MRC Obesity and epigenetics (CR) NIH methylation trajectories in development and ageing (CR) • Future directions ▫ ▫ ▫ ▫ ▫ ▫ Prostate cancer (RM) Insulin resistance/T2D (CR) Air pollution and respiratory phenotypes (JH, PV, PE) UV exposure (JT, AS) Genetical epigenomics (GDS) Other longitudinal studies (MCS, NSHD (1946), Bto20, IMS, APCAPS, Barshi) Acknowledgements • • • • • • • • • • • • George DS Debbie L Sue R Wendy M Beate StP Tom G Adrian S Jon T Luisa Z Nic T Kate T Kate N • Hannah Elliott • Alix Groom