Slides
advertisement
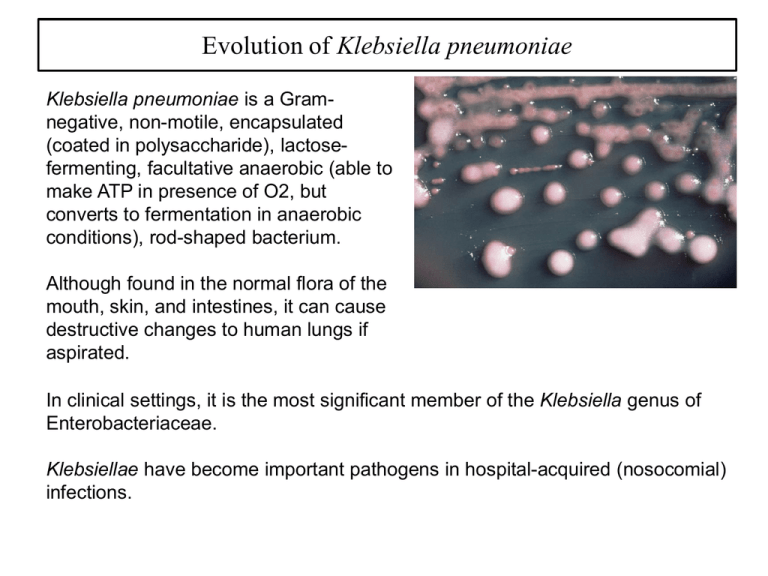
Evolution of Klebsiella pneumoniae Klebsiella pneumoniae is a Gramnegative, non-motile, encapsulated (coated in polysaccharide), lactosefermenting, facultative anaerobic (able to make ATP in presence of O2, but converts to fermentation in anaerobic conditions), rod-shaped bacterium. Although found in the normal flora of the mouth, skin, and intestines, it can cause destructive changes to human lungs if aspirated. In clinical settings, it is the most significant member of the Klebsiella genus of Enterobacteriaceae. Klebsiellae have become important pathogens in hospital-acquired (nosocomial) infections. Klebsiella pneumoniae resistant strains Klebsiella organisms with the ability to produce extended-spectrum blactamases (ESBL) are resistant to many antibiotics, including: • • • • • • • All b-lactams (narrow and broad spectrum inhibitors of cell wall biosynthesis) Aminoglycosides (target protein synthesis in gram-negative bacteria) Fluoroquinolones (broad spectrum disruption of replication) Tetracyclines (broad spectrum disruption of protein synthesis) Chloramphenicol (broad spectrum disruption of protein synthesis) Trimethoprim (DHFR inhibitor) Sulfamethoxazole (Dihydropteroate synthetase inhibitor) One of many carbapenem-resistant Enterobacteriaceae (CRE) is CarbapenemResistant Klebsiella pneumoniae (CRKP). It was first described in North Carolina in 1996; since then CRKP has been identified in 41 states. It is now the most common CRE species encountered within the United States. Over the past 10 years, a progressive increase in CRKP has been seen worldwide. Klebsiella pneumoniae spread CRE spread worldwide SUBURBAN CHICAGO Klebsiella pneumoniae resistant strains The most important mechanism of resistance by CRKP is the production of a carbapenemase enzyme, blakpc. The gene that encodes the blakpc enzyme is carried on a mobile piece of genetic material (a transposon that can jump from chromosome to plasmid and back called Tn4401, which increases the risk for dissemination. CRE can be difficult to detect because some strains that harbor blakpc have minimal inhibitory concentrations (MICs) that are elevated but still within the susceptible range for carbapenems. Because these strains are susceptible to carbapenems, they are not identified as potential clinical or infection control risks using standard susceptibility testing guidelines. Patients with unrecognized CRKP colonization have been reservoirs for transmission during nosocomial outbreaks. KPC Ten variants, KPC-2 through KPC-11 are known, and they are distinguished by one or two amino acid substitutions (KPC-1 was resequenced in 2008 and found to be 100% identical to published sequences of KPC-2). KPC-1 was found in North Carolina, KPC2 in Baltimore and KPC-3 in New York. They have only 45% homology with SME and NMC/IMI enzymes and, unlike them, can be encoded by self-transmissible plasmids. The class A Klebsiella pneumoniae carbapenemase (KPC) is currently the most common carbapenemase. The sequence of KPC is quite different from other BLs G (2a) SHV-1 (2b) TEM-1 (2b) TEM-52 (2be) NMC-A (2f) SME-1 (2f) KPC (2f) KPC (2f) SME-1 (2f) NMC-A (2f) TEM-52 (2be) TEM-1 (2b) SHV-1 (2b) G (2a) The sequence of KPC is quite different from other BLs But its structure is fairly similar G (2a) SHV-1 (2b) TEM-1 (2b) TEM-52 (2be) NMC-A (2f) SME-1 (2f) KPC (2f) G (2a) SHV-1 (2b) TEM-1 (2b) TEM-52 (2be) NMC-A (2f) SME-1 (2f) KPC (2f) KPC SME-1 NMC-A TEM-52 TEM-1 SHV-1 G But its structure is fairly similar Significance Carbapenem-resistant Klebsiella pneumoniae has emerged globally as a multidrug-resistant hospital pathogen for which there are few treatment options. Clinical isolates classified by multilocus sequence typing (ST) as ST258 are the most widespread. The basis for the success of ST258 organisms above and beyond antibiotic resistance is not known, nor is it clear whether infections are caused by a single clone. We used genome sequencing to reveal unexpected genetic diversity among ST258 organisms (thus disproving the single-clone hypothesis) and identified a recombination hotspot that accounts for the majority of divergence—and presumably for serologic variation—among ST258 clinical isolates. Our findings will facilitate the development of new clinical strategies designed to prevent or treat infections caused by multidrug-resistant K. pneumoniae. (more of the same) info on K. pneumonia There is very high mortality (∼30–70%) among patients with bacteremia (blood) or pulmonary (lung) infections caused by carbapenemase-producing K. pneumoniae. The large majority of these isolates are characterized genetically as multilocus sequence type 258 (ST258) and are presumed to be clonally related by descent. Strains of ST258 are resistant to all β-lactam antibiotics and typically contain plasmid-borne genes that encode aminoglycoside-modifying enzymes and chromosomal mutations that confer fluoroquinolone resistance. Carbapenem resistance in ST258 strains is predominantly encoded by K. pneumoniae carbapenemase (KPC), and the gene, blakpc, is located on a transposable element (Tn4401) that is integrated into many different plasmids. Thus, blakpc (along with other plasmid-associated resistance elements) is exchanged readily among K. pneumoniae and other Enterobacteriaceae, a feature key to the spread and high prevalence of multidrug-resistant strains. Multilocus sequence typing Multilocus sequence typing (MLST) is a technique in molecular biology for the typing of multiple loci. The procedure characterizes isolates of microbial species using the DNA sequences of internal fragments of multiple housekeeping genes. Approximately 450-500 bp internal fragments of each gene are used, as these can be accurately sequenced on both strands using an automated DNA sequencer. For each housekeeping gene, the different sequences present within a bacterial species are assigned as distinct alleles and, for each isolate, the alleles at each of the loci define the allelic profile or sequence type (ST). Transmissibility and approach Infections caused by CRKP occur primarily in individuals who have significant clinical risk factors for acquiring these pathogens. These attributes suggest that disease is caused by poor/defective host defense rather than by enhanced bacterial virulence or transmissibility. However, there are no data that bear on these issues, nor is there a clear explanation for the predominance of ST258 globally. We sequenced to closure the genomes of two KPC-producing ST258 K. pneumoniae clinical isolates, and then performed comparative whole-genome sequencing of 83 additional KP clinical isolates obtained from human infections at diverse geographic locations worldwide. Our data provide information that is important for our understanding of the success of ST258 as a human pathogen, extending beyond antibiotic resistance. Transmissibility and approach The two reference ST258 KP clinical isolates, one with mucoid and the other with nonmucoid colony morphology, were obtained in 2010 from patients with urinary tract infections at two separate healthcare institutions in New Jersey. (Note: mucoid indicates a colony showing viscous or sticky growth typical of an organism producing large quantities of a carbohydrate capsule.) The genome sizes were 5,266,518 bp for the isolate named NJST258_1 and 5,293,301 bp for the isolate named NJST258_2, which is similar in length to other K. pneumoniae genomes. Selected K. pneumoniae genomes were compared with the two ST258 genomes, NJST258_1 and NJST258_2. NJST258_1 has 8 putative prophages, 22 insertion sequences (IS), and 2 integrated conjugative elements (ICE). NJST258_2 lacks one of these prophages (phage 6) and has three fewer IS, but the mobile genetic element (MSG) content otherwise is similar to that of NJST258_1. Whether the prophages or other MGEs present in ST258 lineage have contributed to its recent success remains to be determined. K. pneumoniae genome comparisons Plasmid and chromosome-encoded antibiotic resistance NJST258_1 and NJST258_2: Are resistant to β-lactam antibiotics (including carbapenems) and β-lactamase inhibitors (e.g., clavulanate and tazobactam), quinolones (ciprofloxacin and levofloxacin), and aminoglycosides (amikacin and tobramycin). NJST258_1: Is also resistant to trimethoprim-sulfamethoxazole, gentamicin, minocycline, colistin, and polymyxin B, but is susceptible to tigecycline. Many of the antibiotic-resistance determinants are encoded on plasmids in these two strains. Tigecycline Tigecycline is a glycylcycline antibiotic developed by Francis Tally and marketed by Wyeth under the brand name Tygacil. It was given a U.S. Food and Drug Administration (FDA) fast-track approval and was approved on June 17, 2005. It was developed in response to the growing prevalence of antibiotic resistance in bacteria. The NDM-1 and KPC multidrug-resistant Enterobacteriaceae have shown susceptibility to tigecycline. It is structurally similar to the tetracyclines (which is an inhibitor of protein synthesis), but has a substitution at the D-9 position which is believed to confer broad spectrum activity. Tigecycline is bacteriostatic and is a protein synthesis inhibitor by binding to the 30S ribosomal subunit of bacteria and thereby blocking entry of Aminoacyl-tRNA into the A site of the ribosome during prokaryotic translation. Plasmid and chromosome-encoded antibiotic resistance Contain 5 (NJST258_1) vs 3 plasmids (NJST258_2) that vary in size from 10,925–142,768 bp and collectively encode resistance to multiple classes of antibiotics and heavy metals. The 3 plasmids identified in NJST258_2 collectively contained fewer antibiotic and/or heavy metal resistance determinants than did the plasmids in strain NJST258_1. Plasmid and chromosome-encoded antibiotic resistance The quinolone resistance-determining regions in both genomes, including gyrA, gyrB, parC, and parE genes, contained several amino acid replacements that likely explain quinolone resistance in these strains. Taken together, the data indicate that NJST258_1 and NJST258_2 are carbapenem-resistant K. pneumoniae clinical isolates that encode extensive resistance to other classes of antibiotics, resulting in the critical multidrug resistance phenotype. DNA Gyrase Mutations observed in gyrase genes: NJST258_1: S83I and D87G in GyrA, and S80I in ParC NJST258_2: S83I and D87Q in GyrA, and S80I in ParC The list of resistance genes in ST258 is staggering Genome sequencing To gain a better understanding of the emergence and evolution of the ST258 clone, we next performed whole-genome DNA sequencing of 83 additional KP clinical isolates, and mapped the DNA sequence reads to reference strain NJST258_1. Isolates were selected based on the geographic and temporal distributions, KPC variants, and Tn4401 patterns. These clinical isolates were cultured from blood, urine, sputum, or skin of infected individuals or from a rectal swab over a time period encompassing 2003–2012. Isolates were obtained from healthcare facilities at diverse geographic locations in the United States (Florida, Illinois, Pennsylvania, New Jersey, New York, and Texas), Canada, Colombia, and Italy. Seventy-five (89%) of these strains were ST258. The others were ST379 (4), ST512 (4), or ST418 (1), which are all single-locus variants of ST258. Genome sequencing There were 2,436 unique SNPs in the core genome (defined here as all nucleotide positions except those in plasmids and MGE) of all 84 isolates (including NJST258_2) compared with reference isolate NJST238_1. The 84 query isolates differed from NJST258_1 on average by 350 SNPs (range, 116–784 SNPs) in the core genome, indicating that the isolates are closely related. However, in comparison, we previously reported less core genome genetic diversity—an average of ∼50 SNPs—among isolates of the epidemic community-associated MRSA. Of the 2,436 unique SNPs among all isolates, 310 (12.7%) were intergenic, 881 (36.2%) were synonymous, and 1,245 (51.1%) were nonsynonymous. Phylogenetic tree A phylogenetic tree, or evolutionary tree, is a branching diagram or "tree" showing the inferred evolutionary relationships among various biological species or other entities—their phylogeny—based upon similarities and differences in their physical or genetic characteristics. The taxa joined together in the tree are implied to have descended from a common ancestor. Phylogenetic analysis in K. pneumoniae ST258 To estimate genetic relationships, we performed phylogenetic analyses using concatenated SNP nucleotides present in the core genome. All isolates segregated into two distinct subclades. Isolates within clade 1 differed on average by 136 SNPs, and those within clade 2 differed by an average of 82 SNPs. In comparison, the two clades differed from each other by an average of 529 SNPs. 96.3% of the KPC-producing isolates within clade 1 primarily contained plasmids encoding KPC2, whereas 96.2% of those in clade 2 encoded KPC3. The data indicate that the two lineages have had distinct evolutionary histories and thus emerged independently. These findings were unexpected, because the ST258 lineage has been considered to be a single clone or strain. Concatenated SNP nucleotide sequences Initial alignment Seq1 Seq2 Seq3 Seq4 ACTGTAAAGC… ACTGTCCAGC… AGGGTTCAGC… ACGGTGCAGC… Masked alignment Seq1 Seq2 Seq3 Seq4 xCTxxAAxxx… xCTxxCCxxx… xGGxxTCxxx… xCGxxGCxxx… Concatenated SNP alignment Seq1 Seq2 Seq3 Seq4 CTAA CTCC GGTC CGGC Divergence within K. pneumoniae ST258 To better understand the molecular processes that underlie the distinct evolution of clades 1 and 2, we analyzed the distribution of SNPs in specific genes and/or regions of the core genome. Unexpectedly, we found an ∼215-kbp region of the ST258 genome that had a disproportionate number of SNPs. Core phylogeny (-RD) Phylogenetic analysis of the core genome minus this region of divergence (RD) and analysis of the RD alone revealed that the genetic differentiation in the core genomes between the two clades is largely attributed to SNPs in the RD. RD chromosomal segment RD phylogeny Capsule polysaccharide Capsule polysaccharide (CPS) is a well-known contributor to the success of K. pneumoniae as a pathogen, in part because the variation in genes that encode enzymes involved in CPS synthesis results in serologic diversity. Notably, the CPS gene island, which encodes the capsule K-antigen in K. pneumoniae, is located within the RD. Currently, more than 78 different Kserotypes have been identified, some of which are significantly associated with serious human infections. Unexpectedly, we identified two distinct cps gene clusters in the genomes of the ST258 clinical isolates (ST258 cps 1 in clade 1 and cps 2 in clade 2). The genetic differences between these two cps clusters are primarily responsible for the segregation of ST258 isolates into two distinct clades. Capsule polysaccharide Concluding remarks CRE—most notably ST258 K. pneumoniae—now are classified as an “Urgent Threat” by the CDC. Although the absolute number of estimated deaths caused by CRE is relatively low (in the United States 610 deaths annually versus 11,000 caused by MRSA and 14,000 caused by C. difficile), the potential exists for transfer of multidrug resistance to Gram-negative community-associated pathogens such as E. coli. As suggested by the CDC report on antibiotic resistance threats, a multi-tiered approach is needed to prevent or moderate the threat posed by CRE. Such an approach includes understanding the evolutionary genomic history of the microbe as a means to gain new insight into its success as a human pathogen, as we have done herein. Unexpectedly, we discovered that ST258 is comprised of two distinct lineages or clades rather than being a single clone as previously suggested, which are differentiated largely by an RD that encodes CPS biosynthesis machinery. Thus, independent acquisition of genetic material by an ancestral ST258 clone (rather than within-ST258 diversification) is the primary basis for segregation of into two clades. Concluding remarks CRE—most notably ST258 K. pneumoniae—now are classified as an “Urgent Threat” by the CDC. Although the absolute number of estimated deaths caused by CRE is relatively low (in the United States 610 deaths annually versus 11,000 caused by MRSA and 14,000 caused by C. difficile), the potential exists for transfer of multidrug resistance to Gram-negative community-associated pathogens such as E. coli. As suggested by the CDC report on antibiotic resistance threats, a multi-tiered approach is needed to prevent or moderate the threat posed by CRE. Such an approach includes understanding the evolutionary genomic history of the microbe as a means to gain new insight into its success as a human pathogen, as we have done herein. Unexpectedly, we discovered that ST258 is comprised of two distinct lineages or clades rather than being a single clone as previously suggested, which are differentiated largely by an RD that encodes CPS biosynthesis machinery. Thus, independent acquisition of genetic material by an ancestral ST258 clone (rather than within-ST258 diversification) is the primary basis for segregation of into two clades.