Group 6 Final Presentation Powerpoint
advertisement

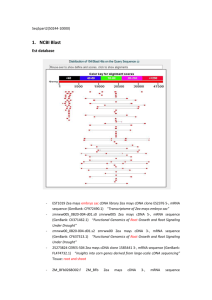
Bikash Shakya Emma Lang Jorge Diaz BLASTx entire sequence against genomes. RepeatMasker 55.47% repetitive sequences 82.5% retroelements 13.0% DNA transposons EMBOSS explorer 74 CpG islands 54 inverted repeats 9 plant GENE PREDICTION Masked sequence Unmasked sequence GeneMark GeneMark 12 genes 27 genes FGENESH FGENESH 10 genes 28 genes BLASTx 7 most promising genes Bases: •START & STOP codons •High GC content •No repeats •Good E-value •Proper splice sites •Both program agreed •No mobile elements GENE I: Zea mays uncharacterized protein LOC100194332 Both programs predicted the exact same 3 exons RNA Evidence BLAST search in the refseq_rna database Zea mays uncharacterized LOC100194332 (LOC100194332), mRNA (cDNA) Identity:100% E-value:0 Sequence alignment with the translated sequences GENE I Perfect match Identity:99% E-value:0.0. EST data covered both exons 1 & 2 except 114 bases GENE I Protein function • Conserved domain: Myb DNA binding • Predicted to be a MYB related transcription factor • Myb proteins bind to DNA and regulate gene expression 6 exons 241 amino acids membrane protein with 7 transmembrane helices sugar efflux transporter Image from: http://bp.nuap.nagoya-u.ac.jp 99% match to “Zea mays seven-transmembranedomain protein 1” (LOC100284352) mRNA (cDNA) EST data covered all of exons 1, 2, 3, and 4 plus beginning of exon 5 ◦ All EST sequences used had 98-99% identity with gene II conserved domain: MtN3_slv Sugar efflux transporter Involved in seed and pollen development Starch branching enzyme I from rice. 1 exon 899 amino acids Soluble protein 1,4-alpha-glucanbranching enzyme 3/ starch branching enzyme 3 Matched orthologs in 5 other plant genomes. Image from: http://pdb.rcsb.org 99% match to “Zea mays starch branching enzyme III (sbe3)” mRNA (not cDNA) EST data covered almost all of gene III (1 gap) (intron?) ◦ All EST sequences used had 99%-100% identity with gene III Segment without EST data aligns to starch branching enzyme III in A. thaliana – not an intron conserved domains for 1,4-alpha-glucanbranching enzyme top HHpred result was starch branching enzyme 1 in rice (e-value: 2e-128) These enzymes catalyze the formation of the alpha-1,6-glucosidic linkages in starch. 5 exons 583 amino acids Membrane protein with 10 trans-membrane helices Amino acid transporter Matched orthologs in wheat and sorghum genomes. 96% match to “Zea mays LOC100193963 (si486073c04), mRNA” (E=0.00) (not cDNA) Other good match was to “XM_002455881.1Sorghum bicolor hypothetical protein, mRNA” (94%, E=0.0) EST best matches: ◦ ZM_BFc Zea mays cDNA clone ZM_BFc0171C07 5‘ (95%, E=0.0) ◦ ZM_BFc Zea mays cDNA clone ZM_BFc0038P24 5‘ (96%, E= 2e-158) EST data also have two gaps. Conserved domains: ◦ NCBI BlastX ◦ InterProScan