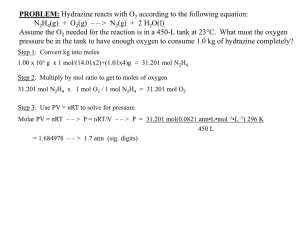
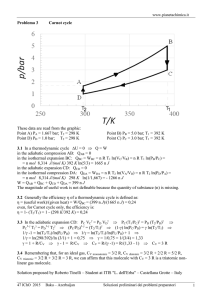
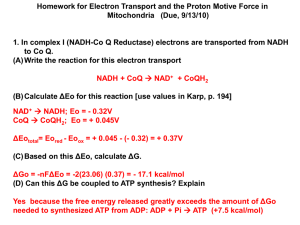
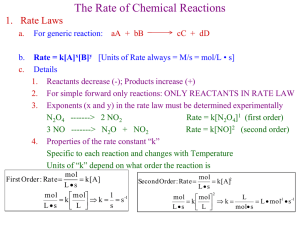
CD (Cognitive Dissonance) and MD
advertisement

CD and MD What’s my problem with MD? 1. Its development has been manifestly unscientific 2. Its answers (numbers, trajectories, minima) are as unreliable (or more) than simpler methods 3. Yet its manifest societal advantages- “physics”, movies, CPU time, complexity, jargon- lead to cognitive dissonance (hopeful thinking) concerning its actual value to drug discovery CD: Cognitive Dissonance Wikipedia:Cognitive dissonance theory explains human behavior by positing that people have a bias to seek consonance between their expectations and reality. According to Festinger, people engage in a process he termed "dissonance reduction," which can be achieved in one of three ways: lowering the importance of one of the discordant factors, adding consonant elements, or changing one of the dissonant factors. This bias sheds light on otherwise puzzling, irrational, and even destructive behavior. Lowering importanceAdding consonanceChanging the dissonance- Actually agreeing (numerically) with experiment “It’s an idea generator” Reparameterizing (+ Effort Justification Paradigm) AM I CD? • Came from Barry’s Lab (the Great PB MD Wars) • Don’t sell MD (perhaps I’m jealous) Why should you believe me? -Don’t write/ need grants -Don’t need tenure -PB is not a significant OE income stream -Been observing MD for > 25 years -I hired an MD guy (who I sent to China!) -I manifestly want this to be a better industry Also.. • • • • • • The fastest PB- DelPhi, ZAP The fastest surfacing algorithms- GRASP, ZAP The fastest 3D shape alignment- ROCS, FastROCS The fastest conformer generator- OMEGA The fastest, non-stochastic docker- FRED The fastest (accurate) Surface Area, RMSD, AM1, protein pka, proton placement.. • If I wanted to do MD, mine would rock • I believe the effort/reward ratio is (way) too low How Galileo Transformed Science Think something up 1. Resolution See if it matches available evidence 2. Demonstration 1. Experiment Think of a new experiment to test it (to differentiate from old theories) A Galilean Value Scale for Experiments • Retrospective Data that shapes the theory – MD, Most of molecular modeling, economics • Prospective without Controls – Rich Friesner, Xavier Barril Better • Unanticipated Retrospective Data – SAMPL solvation energies • Prospective designed with NULL model Controls – Bertrand Garcia Moreno, protein pKa Collective – Lyall Isaacs, SAMPL host-guest • Prospective to distinguish from Best-of-Class Controls – Nobody A Galilean Value Scale for Experiments • Retrospective Data that shapes the theory – MD, Most of molecular modeling, economics Vast Majority • Prospective without Controls – Rich Friesner, Xavier Barril • Unanticipated Retrospective Data – SAMPL solvation energies Better • Prospective designed with NULL model Controls – Lyall Isaacs, SAMPL host-guest – Bertrand Garcia Moreno, protein pKa Collective • Prospective to distinguish against Best-of-Class Controls – Nobody Prospective Without Controls • Surgeons coming up with new procedures – Osteoarthritis & Arthroscopic knee surgery • US Foreign policy – Just do something, claim success when it works, bury it when it doesn’t • Anecdotal stories – The “hot hand” phenomena – I did “X”, it worked. I did “X”, it worked Two chief fallacies (i) Fallacy of Composition -What else did you actually do (ii) Fallacy of Selection -File Drawer effect (False Positives) -Parameterization (implicit or explicit) to the result (False Negatives) Fallacy of Composition • Method X, e.g. MD, is but one part of a multipart process (filtering, chemists inspection, database bias)- success is claimed for X alone • The same procedure with X replaced with a different method is never done/ presented Example of Composition Error • We predicted affinity with MM/QM and “It Worked” • Was QM getting you anything? • Did you do MM with QM-level charges, multipoles? MM alone? A scoring function? Example of Composition Error • We used a polarizable force field and got these results for the (SAMPL4) host-guest systems. “It Worked”, so polarization worked. • Did you also try it without polarization? With better quality charges? With equivalent CPU time but without polarization (more sampling)? Example of Composition Error • We ran MD for a bit, looked at how the ligands wiggled and designed six drugs (Christopher Bayly & others at Merck Frosst) • Did you compare to MM? To other simple heuristics? Without any chemists input? • It’s not “Science” until someone else does it Fallacy of Selection: The Tanimoto of TruthTM An Event Happened Reality Predictions An Event Didn’t 0 1 1 0 1 0 0 1 0 1 1 1 1 0 0 1 0 1 0 0 ToT = Events that happened and were predicted Events predicted or happened The TheTanimoto Tanimotoof ofTruth Truth An Event Happened Reality Predictions An Event Didn’t 0 1 1 0 1 0 0 1 0 1 1 1 1 0 0 1 0 1 0 0 Published Especially by Academia The TheTanimoto Tanimotoof ofTruth Truth An Event Happened Reality Predictions An Event Didn’t 0 1 1 0 1 0 0 1 0 1 1 1 1 0 0 1 0 1 0 0 “File Drawer” False Positives Especially by Industry The TheTanimoto Tanimotoof ofTruth Truth An Event Happened Reality Predictions An Event Didn’t 0 1 1 0 1 0 0 1 0 1 1 1 1 0 0 1 0 1 0 0 False Negatives- Parameterize till publishable Especially by Academia The TheTanimoto Tanimotoof ofTruth Truth An Event Happened Reality Predictions An Event Didn’t 0 1 1 0 1 0 0 1 0 1 1 1 1 0 0 1 0 1 0 0 True Negatives- Not sexy, “Hempel’s Ravens” Largely ignored by Academia & Industry The TheTanimoto Tanimotoof ofTruth Truth • “Similarity” methods, Docking, Machine Learning • All are judged by some kind of ToT • Quantification for MD ‘events’? Never. • MD is mostly uncontrolled, anecdotal & unscientific Psychology, Philosophy, Social Dynamics Underlying Physics, Examination of Successes Molecular Dynamics: Types of Applications 1) Global sampling- thermodynamic averages -FEP etc. Absolute or Relative Energies 2) Simulate time evolution (movies) -D.E. Shaw, Vijay Pande- Mechanism 3) Local sampling (thermally accessible barriers) -Bayly & co., WaterMap, MM/PBSA. Qualitative Assessment Thermodynamic energies and Fables of Physics “We all know that if we had the perfect force field and simulated for an infinite time, we’d get the right answer”- Woody Sherman, ACS San Francisco, March 24th, 2010 1) pKa, Tautomers 2) Finite temperature, MD & Stat Mech 3) Ergoticity? 4) The illusion of a ‘perfect” ForceField (that ≠ QM) Typical FF Thinking: Polarization • Polarization is tricky • But it makes dipoles bigger, e.g. water – 1.85D (vacuum) 2.5~2.6D (condensed phase) • So therefore increase charges by ~15% – E.g. use HF-6-31G* • Now molecules are roughly correct Polarization of Dipoles -|+ E 0 -|+ E 0 -|+ D- + -|+ + - + + + Favorable +|Unfavorable - -|+ - Epol - -|+ D - - - - - - +|- - - Epol Scaling vs Polarization Alignment Scaling Charges Polarization Favorable Unfavorable Lowers Energy Lowers Energy Raises Energy Lowers Energy Scaling dipoles can only be accurate on average (with parameterization) not locally! Ah, but then there’s AMOEBA EPIC Quantum mechanics (“PB”!) 5 PID AMOEBA 5 -75 -50 -25 0 25 50 4 3 5 -10 -75 -50 -25 0 25 50 4 3 0 + 2 -10 -75 -50 -25 0 25 50 4 3 + 2 0 -10 0 + 2 -75 -75 1 1 0 C H C 0 + -3 C H C H 0 -1 + -2 -3 -4 -4 -5 -5 -5 -4 -3 -2 -1 0 1 2 3 4 5 X (angstrom) -75 10 Y (angstrom) Y (angstrom) Y (angstrom) -1 -2 1 10 H C H C H 10 -1 + -2 -3 -4 -5 -4 -3 -2 -1 0 X (angstrom) 1 2 3 4 5 -5 -5 -4 -3 -2 -1 0 1 2 3 4 5 X (angstrom) (Jean-Francois Truchon) Kim Sharp: JF Applications: cation-p 0 Acetylcholinesterase -10 -20 O O N+ NH -30 B3LYP Li+ B3LYP Na+ B3LYP K+ polarizable (DRESP, P2E) non-polarizable (RESP) -40 -50 -60 -70 Electrostatic i 2.0 2.5 3.0 3.5 4.0 4.5 Cation / benzene distance (angstrom) JF Hydrogen Bonds: Formamide dimer “ Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations” A. V. Morozov, T. Kortemme, K. Tsemekhman and D. Baker, PNAS, Volume 101, page 6946, 2004. Method δ(HA) ψ ϴ X DFT 1.94 112.34 159.43 -177.51 MP2 1.97 110.49 155.33 -179.49 HF 2.10 138.16 170.94 -179.54 CHARMM27 1.82 170.25 170.83 -106.83 OPLS-AA 1.75 165.04 175.61 145.12 MM3-2000 1.98 121.16 161.07 149.63 PDB 1.93 115.00 175.00 175.00 Geometry optimizations starting from the Baker MP2 minimum Geometry optimizations starting from the Baker MP2 minimum Geometry optimizations starting from the second MP2 minimum Geometry optimizations starting from the second MP2 minimum Ah, but then there’s AMOEBA R(O..H) (Å) 0 E_ele (kcal/mol) 1.9 2.2 2.5 2.8 3.1 3.4 3.7 -3 QM* *CCSD/aug-cc-pVTZ Pt. Charge Pt. Octupole -6 -9 Fitting to the electron density Denny Elking, Tom Darden Model Point Monopole Point Dipole Point Quadrupole Point Octupole Exponential Monopole Exponential Dipole Exponential Quadrupole Exponential Octupole CCSD/aug-cc-pVTZ Electrostatic Energy (kcal/mol) -4.33 -5.81 -6.36 -6.31 -7.68 -8.32 -8.52 -8.18 -8.23 Or…… Model Point Monopole Point Dipole Point Quadrupole Point Octupole Exponential Monopole Exponential Dipole Exponential Quadrupole Exponential Octupole CCSD/aug-cc-pVTZ Electrostatic Energy (kcal/mol) -4.33 -5.81 -6.36 -6.31 -7.68 -8.32 -8.52 -8.18 -8.23 Increase Dipole from 1.85D to 2.56D Details, Details.. 1) Just incorporate Volume Terms (PB) 2) And all those other terms: - Exchange interactions - VdW anisotropy - pKa & Tautomers - Cross-terms between valence and non-bonded - Three (N) body terms…. Eventually it’ll be right! Woody’ll be right. Inconceivable it can’t ever be right. (Wolynes) Concrete MD Examples • Binding Energies- Shirts - Also Solvation (Simpler system) • Protein Trajectories- Shaw - Also Peptides (Simpler systems) • “Minimization” – Shoichet - Is a simple system FKBP-12 Unanticipated Retrospective Data? FKBP-12: Shirts et al (Thesis) -5 -15 -14 -13 -12 -11 -10 -9 -8 -7 -6 Simulation (kcals/mol) -7 -8 -9 -10 -11 -12 -13 -14 -15 Experiment (kcals/mol) FKBP-12 Again FKBP-12: Shirts et al (My plot) -5 -14 -13 -12 -11 -10 -9 -8 -7 -6 -5 -6 Simulations (Kcals/Mol) -7 -8 -9 -10 -11 -12 -13 -14 Experiment (Kcals/Mol) FKBP-12 Yet Again Retrospective Data that shapes the theory FKBP-12: Shirts et al (My plot: No Long Range Correction) -2 -14 -13 -12 -11 -10 -9 -8 -7 -6 -5 -4 -3 -2 -3 -4 Simulations (Kcals/Mol) -5 -6 -7 -8 -9 -10 -11 -12 -13 -14 Experiment (Kcals/Mol) Contributions to Affinity Desolvation Entropy VdW Discrete Waters Coulombic Polarization Buried Area Shape Correlations to Affinity Buried Area Entropy VdW Polarization Coulombic Discrete Waters Desolvation Electrostatics E.g. VdW Train on 17 HIV-1 Protease Inhibitors 1) Minimization (MM2X) 2) pIC50=-0.15*Einter-8.1 Prospectively used on 16 more E.g. Coulombic • Urokinase Brown & Muchmore, JCIM, 2007, (47) 4 Coulombic Interaction E.g. Buried Area “Fast and Accurate Predictions of Binding Free Energies using MM-PBSA and MM-GBSA”Rastelli, G., Del Rio, A.,Degliesposti,G., Sgobba, M. J. Comp. Chem. Vol 31, #4, pg 797-810 Buried Area MM-PBSA 0 0 -15 -10 -5 0 -10 DHFR -20 -30 -40 y = 2.339x - 11.308 R² = 0.8494 -50 -60 Experimental Binding (kcal/mol) -20 Buried Area Energy (kcal/mol) Predicted Binding (kcal/mol) -20 -15 -10 -5 0 -1 -2 -3 -4 y = 0.3181x - 0.7159-5 R² = 0.7474 -6 -7 Expt. Binding (kcal/mol) My observation over 20 years • For congeneric series, something basic often correlates, sometime well (VdW, Coulombic) • For non-congeneric usually nothing works • If something works for non-congenerics, it’s usually something basic (mass, buried area) Simpler System: Solvation # Compounds MD RMSE kCal/mol PB RMSE kCal/mol 1.76 (Me) SAMPL0 17 1.35 (Vijay) SAMPL1 56 3.6 (Mobley) 2.2 (Me) SAMPL2 40 2.4 (Jay) 2.1 (Ben) SAMPL4: 50 Solvation Energies My PB Method Best MD QM + Specific Group-wise Parameterization Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs- D. E. Shaw 1) Where they bind - Confirmed by mutagenesis 2) A surprise in how they bind -pi-charge interactions -not charge-charge 3) Cause of allostery: (i) Charge (ii) Binding pocket width -Confirmed by synthesis IMHO 1) Where they bind 1) Docking with Glide did almost - Confirmed by mutagenesis as well. Confirmation is WEAK. 2) How they bind -pi-charge interactions -not charge-charge 2) THIS IS NOT A SURPRISE! 3) Cause of allostery: (i) Charge (ii) Binding pocket width -Confirmed by synthesis 3) (i) Already known & follows charge multiplicity exactly. (ii) –ONE CMPD (better than most!) Also.. • Local ionizable residues never (de)protonate – Binding +3 ligands • NMS was modeled, not simulated • Experimental errors claimed are <0.1 kcal in vivo Simpler Story- Peptides • Poly-Ala propensities (2010) – Have to modify FF to get helicity right • Side-chain conformation preferences (2012) – Little agreement between force-fields – Poor agreement with crystals (2013) • H-bond geometries (2005) – Flawed Baker study • Beta-hairpin simulations (2012) – Little agreement between force-fields Simple System: ShoichetRelative binding energies in a cavity A signal! Poses selected, not found, so is this dynamics or minimization? Maybe not! NULL MODELS RMSE from Phenol = 2.5 kcal/mol RMSE from from Catechol = 1.1 kcal/mol RMSE of the “NULL” hypothesis = 1.2 kcal/mol From “closest” Phenol|Catechol = 0.8 kcal/mol One, Inescapable, Conclusion • We cannot calculate the energies of protein microstates with any accuracy • It is unclear even how bad we are • Even ranking must be suspect Of Dubious Value • Ranking Ligands, Absolute or Relative • Flexible Docking • Protein folding to atomic resolution • Evaluating unfolded states • Excursions from the crystal structure So how can we fold (small) proteins? • Luck- are small proteins self-selectingly robust? • Some parameterization (Shaw) • Stability of kinetic pathways might be more robust than energetics suggest (Pande) ? But what’s the alternative? • To Local Minimization – Sample (MC, Low Mode etc) and minimize • To Energy evaluation – Exhaustively sample and minimize • To time evolution – Elastic network? Low mode dynamics? – Run MD! Experiments I Wish Were Done • Protein Crystallography – Predict the room temperature density • Small molecule NMR – Predict the dominant low energy conformer • Protein Electrostatics – Predict potentials in the active site • Host-guest systems – Binding energies, salt effects And how I wish they were done: Maximal Disinformation Testing 1. FIRST calculate for two or more methods, e.g. polarization vs static, PB vs MD, MD vs MM 2. Prospectively measure those systems that most distinguish methods- mutual disinformation 3. Adapt theories- no one’s perfect! 4. Repeat steps 1,2 & 3 5. Does a prediction ‘gap’ persist? E.g. Kepler vs Epicycles. Final Thoughts • I’d love MD to work! Make my job easier • It doesn’t. At least not as advertised/ believed • It’s nature (“physics”, big calculations, movies) leads to overconfidence • Until a more scientific approach is adopted it’s unlikely to get better. GPUs won’t save MD • What’s needed is Maximal Disinformation Testing & Model systems