lsum2013_sakyuz
advertisement

COMPARISON OF FTIR SPECTRA OF
BIOMOLECULES ACQUIRED USING
SYNCHROTRON IR AND THERMAL IR SOURCES
Sevim Akyüz
Physics Department, Istanbul Kültür University, Ataköy
Campus, Bakirköy, 34156, Istanbul, Turkey
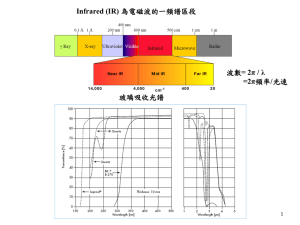
FTIR spectroscopy is a powerful tool for qualitative and quantitative analysis
of molecular structures. It has the ability to characterize different samples
(for instance, organic and inorganic materials, or crystalline and amorphous
materials) in the same time. Any alterations in the molecular composition or
molecular orientation can be detected.
On the other hand IR micro-spectrometry is preferable for analyzing
inhomogeneous structures. For investigation of biological molecules or
cultural heritage metarials, that have inhomogeneus structures, generally IR
micro-spectroscopy is used which has ability to monitor the chemical
composition point by point, using the inherent spectral properties of the
components.
But the spatial resolution for conventional IR micro-spectrometry
measurements is limited.
Synchrotron radiation based Fourier-transform infrared (S-FTIR) microspectroscopy is a new-developed technique, which combines the advantages of
FTIR spectroscopy and synchrotron light source. Synchrotron light source is used
as external IR source.
Conventional IR spectrometers have thermal sources (Globar or Nents Glower), they
produce radiant energy based on black body radiation. They produce IR spectrum
from a filament heated to 1000 to 2000 K. These seramic sources are physically large
(at least several millimeters), and typically radiate in all directions. The optics of the
FTIR bench collects and collimates the IR radiation, passes it through the
interferometer, and then on to the IR microscope where the modulated IR light is
focused to a small spot on a sample. Finally, the IR radiation, reflected or transmitted
from the sample is collected, focused onto an appropriate infrared detector, and
processed by a computer via a Fourier transform to produce an infrared spectrum.
The thermal emission sources can be focused to 75–100 μm
diameter with an IR microscope. In order to measure
something smaller, we must mask away part of the incoming
light, significantly reducing the signal strength.
The spatial resolution for IR-MSP measurements is limited in
practice by two factors.
The first factor is the size of the aperture used to limit the size
of the image that is projected through the IR microscope onto the
detector. In order to resolve individual cells, or components
thereof, apertures as small as 10 m are desirable.
However, the low intensity of light passing such a small
aperture seriously restricts the ability to collect an absorption
spectrum at such high spatial resolution.
The second factor is a diffraction effect that becomes significant when
the size of the aperture approaches the wavelength of light utilized.
A true point source could be focused to a diffraction-limited spot
size, which is approximately the wavelength of the light.
This can be achieved with a synchrotron IR.
For mid-IR and longer wavelengths, the effective source size for a
synchrotron light source is diffraction-limited spot size.
In other words, it is very close to an ideal point source. The beam is
focused to a spot with a diameter ≤10 μm, which is smaller than a
typical mammalian cell, and provides thousand times the brightness
of conventional IR sources.
Synchrotron as an IR source makes a big improvement. Synchrotron radiation
is a high-brightness infrared source, which is well suited to spectroscopy. The
spectral range,which extends from about 2.5 to 20 micron, so-called midinfrared region, spans most of the vibrational mode frequencies necessary to a
detailed identification of functional chemical groups. The primary advantage of
synchrotron infrared radiation is its brightness.
The brightness of synchrotron light is defined as the photon flux or
power emitted per source area and solid angle.
It is important to note that synchrotron radiation does not provide a much
higher photon flux than a conventional IR source (such as a globar).
The crucial factor is that the effective synchrotron source size is quite small,
i.e. on the order of 100 m or less. Thus, the light is emitted into a narrow
range of angles, and the resulting brightness is vastly increased.
Synchrotron Source Advantages
Higher spatial resolution
Brightness
Better signal-to-noise ratio
(1000 x)
Faster collection
+
Broadband
Spectroscopy
Heating at sample is negligable
Lateral resolution is limited by diffraction; in confocal
configuration it is l/2
The ultra-brightness and high spatial resolution of
synchrotron light source ensure S-FTIR micro-spectroscopy
is capable of analyzing small samples or small sample areas
that conventional FTIR cannot do.
Due to high brightness, infrared light from a synchrotron
can be easily collimated and/or focused to diffraction
limited spot sizes (~1-10 μm), allowing high spatial
resolution and high spectral resolution.
IR spectra of single red blood cell; recorded with synchrotron IR (red)
and with conventional IR spectrometers1.
1- D. McNaughton, Australian Biochemist,36(1), (2005) 55-58
IR spectra obtained on the same individual cell (HELA cell), using either the
synchrotron beam (a), or thermal source (globar), with an aperture of 6 x 6 (µm)2.
Taken from: P. Dumas, L.M. Miller and M.J. Tobin, Acta Physica Polonica A, 115
(2009) 446-454
Synchrotron-based FTIR spectrum in plant seed endosperm tissue (wheat) (pixel
size: 10m×10m). Taken from ref 2.
Ca. 1720-1500 cm-1 = protein (amide I and amide II bands)
ca. 1185–800 cm−1 = total carbohydrate
ca.1065–950cm−1 = non-structural carbohydrate (starch) region
2- P. Yu, H.C. Block, K. Doiron, Spectrochimica Acta Part A 71 (2009) 1837–1844
INTERACTION OF 24EPIBRASSINOLIDE WITH CULTURED
PROSTATE CANCER CELLS
(DU145, PC3) AND EPITHELIAL CELLS
(PNT1A): AN FT-IR SPECTROSCOPIC
STUDY
Sevim Akyuz1, Tanil Akyuz1, Pinar Obakan2, E. Damla
Arisan2, Narcin Palavan-Unsal2
1Istanbul
Kultur University, Department of Physics, Atakoy Campus,
34156 Bakirkoy, Istanbul, Turkey, e-mail: s.akyuz@iku.edu.tr
2Department
of Molecular Biology and Genetics, Atakoy Campus,
34156 Bakirkoy, Istanbul, Turkey
Brassinosteroids (BRs), belong a class of polyhydroxysteroids,
are growth-promoting plant steroids that possess various types
of regulator activity in plants. In addition to stimulating growth,
BRs have an anti-stress effect on plants. They have the ability
to overcome both abiotic and biotic stress in plants [3].
Recently it was shown that natural brassinosteroids can inhibit
the growth of various cancer cell lines at micromolar
concentrations, despite having minimal effects on normal cells.
Therefore brassinosteroids are promising leads for potential
anticancer drugs. 24-Epibrassinolide (C28H48O6) is a
brassinosteroid [4].
[3] C. Brosa, Crit. Rev. Biochem. Mol. Biol., 34 (1999), 339–358.
[4] J. Steigerova, J. Oklestkova, M. Levkova, L. Rarova,Z. Kolar, M. Strnad,
Chem-Biol Interact 188 (2010), 487–496.
In this study the interaction of cultured prostate cancer cells (DU145 and
PC3) with 24-epibrassinolide, a promising drug candidate in cancer
therapy, have been investigated using Fourier transform infrared
spectroscopy (FTIR) in order to investigate the alterations of intracellular
composition and conformation, induced by epibrassinolide. We also
investigated the effects of epibrassinolide on prostate epithelial cells
(PNT1A).
24-epibrassinolide
C28H48O6
An infrared spectrum of a biological sample is composed of characteristic
absorption bands originating from all molecules it contains, such as
proteins, lipids, nucleic acids, and others.
FTIR spectroscopy, one of the few spectroscopic techniques that can
detect changes in many different biomolecules simultaneously and in situ,
has proven to be a useful tool for the detection of molecular changes
directly in the cells, tissues.
In this study we carefully investigated protein
structure by analysing amide I, amide II and amide III
bands, DNA structure by analysing phosphodiester
group of PO2- stretching bands around 1250-1080 cm-1
and DNA bands of the backbone chain around 830-880
cm-1 regions
Amide I
mode
Protein conformation
Amide I mode is primary a C=O stretching motion of amide groups,
couples slightly with C-N stretching, N-H bending and CCN deformation
motions [5,6].
Amide II mode is N-H in plane bending motion coupled with C-N
stretching [5].
The Amide III mode is mainly in-phase combination of NH in-plane
bending and CN stretching motions. It has minor contributions from CC
stretching and CO bending [5]
Due to the overlapping of protein amide component peaks, second derivative
profiles of the absorption spectrum was used to identfy amide I, amide II and
amide III component peaks, then multicomponent peaks fitting with Gaussian
function was used to quantify the multicomponent peak area in amide regions.
W.K. Surewicz, H.H. Mantsch, “Infrared Absorption Methods for Examining Protein
Structure” in “Spectroscopic Methods for Determining Protein Structure in Solution”, (Ed. H.A.
Havel),VCA pub. Inc, 1996, NY.
[6] A. Barth, Biochim. Biopys. Acta 1767 (2007) 1073.
[5]
Cultured
prostate
cancer cells
Amide I and amide II
regions of protein
structure
Water bands are
subtracted
Cancer cells
containing 25 M
epibrassinolide,
a promising drug
candidate in
cancer therapy
Amide I and
amide II
regions of
protein
structure
Comparison of the second derivative profiles of the IR absorption spectra of DU145
prostate cell line containing 0 and 25 M Epibrassinolide, after subtracting water bands.
In addition to alterations in proteins conformation,
significant spectral changes were also observed at the
intensities of characteristic DNA/RNA bands of prostate
cancer cells.
Particularly asymmetric phosphate (PO2-) stretching mode
of phosphodiester backbone of nucleic acids is affected by
the presence of epibrassinolide. It was found that the
hydrogen bonding of the phosphodiester groups of
nucleic acids decreases with the presence of
epibrassinolide, which indicates alterations in DNA
packing condition.
a PO 2
C
1485-1010 cm-1 region of the IR spectra of prostate cancer cell line DU145,
with and without epibrassinolide
ca.1401 cm-1 cytidine; ca.1239 cm-1 PO2- asimetric stretching; ca1087 cm-1 PO2symmetric stretching vibrations.
Band component analysis of the 1340-1020 cm-1 region of the IR
spectrum of prostate cancer cell line DU145
The 1222 cm-1 and 1244 cm-1 bands of LNCaP cell lines are assigned to strongly
hydrogen bonded and weakly-hydrogen bonded phosphodiester groups of nucleic
acids. The integrated intensity ratio of these bands for LNCaP ((I1222 / I1244) is found
to decrease by the addition of EBR,
DU145 +EBR
Band component analysis of the 12800-1020 cm-1 region of the IR
spectrum of prostate cancer cell line DU145 containing 25M
epibrassinolide
When we compared the band components of the PO2- asymmetric stretching
mode of the phosphodiester group of DNA we sow hydrogen bonding of the
phosphodiester groups of nucleic acids decreases with the presence of EBR,
which indicates alterations in DNA packing condition
PC3 is another prostate cancer cell line
In a previous study, It was also shown that in B to A transition B-DNA marker band
1228 (*) shifts to 1240 (**)
{D.K. Jangir, G. Tyagi, R. Mehrotra, S. Kundu, J. Mol Struct 869 (2010) 126-129}
Epithelial cells {Not-cancereous; normal cell line}
Band component analysis of the FTIR spectrum of the 1485-1010 cm-1 region of
epithelial cells (PNT1A)
We did not see the same significant alteration in phosphodoester bands of
DNA by the presence of EBR in epithelial cells (PNT1A)
EPITHELIAL CELLS (PNT1A):
We did not see the same significant alteration in epithelial cells (PNT1A) by the
presence of epibrassinolide
1000-700 cm-1 region of prostate cancer cell line DU145
B-DNA
A-DNA
836 cm-1 is a marker band of B-DNA; 869 is a marker band of A-DNA.
Presence of epibrassinolide causes partial BA transition of DNA in DU 145 cell
line.
.
In order to reveal the data embedded in the molecular structure of prostate cancer
cells containing 25M epibrassinolide in comparison with those of the control
group, we performed a chemometric method, principle component analysis (PCA).
Principle component analysis was performed on IR spectra of control group and
EBR containing prostate cancer cells, after subtracting water bands using The
Unscrambler CAMO software.
PCA is a multivariate analysis technique that reduces data matrix to their lowest
orthogonal space, and found to be very informative to obtain hidden data
structures [8]. Mathematically, PCA is based on an eigenvector decomposition of
the covariance matrix of the variables in a data set. Given a data matrix X with m
rows of samples and n columns of variables, the covariance matrix of X is defined
as:
{Cov}(X) = (XTX) / m-1
Here XT is transpose matrix of data matrix X and m is the number of samples.
The result of the PCA procedure is a decomposition of the data matrix X into
principal components called score and loading vectors. As matrix notation we can
write data matrix X:
X = T PT + E
Here T is the score matrix, PT is the transpose of P loading matrix and E is the
residual matrix. The score matrix (T) is related to the sample contributions, and
the loadings matrix (P) is related to the variables contributions.
The score and loading matrixes contain information on how the samples and
variables, respectively, relate to each other.
In the present study, X matrix contains IR spectra of control group (DU145
prostate cell line) and 25 M Epibrassinolide containing DU145 cell line
(after subtracted water bands). Every sample spectrum is an average of 5
replicate spectra, 6 control, and 7 epibrassinolide containing samples
were analyzed.
The components PC1 and
PC2 (Figure) show all
contribution to the variance
in concentrations (PC1 = 95
%, PC2 = 5%), which are
enough to separate the
samples.
In order to reveal the similarities and dissimilarities between control
group prostate cancer cell and epibrassionilide containing cells,
Hierarchical Average linkage cluster analysis were performed on the
1800-600 cm-1 region of the vector normalized IR spectra (water bands
were subtracted) using Euclidean distance.
The dendrongram is as follows:
CONCLUSIONS
IR spectroscopy has proven to be a a powerfull tool for the detection of molecular
changes in the cells.
Significant spectral changes were observed at the intensities and frequencies of
characteristic protein and DNA vibrational bands of prostate cell lines. Hydrogen
bonding of the phosphodiester groups of nucleic acids and phospholipids decreases
with the presence of EBR, which indicates alterations in DNA packing condition.
Presence of epibrassinolide causes partial B A transition of DNA
We also observed alterations in amide I and II bands which may indicate
alterations of the secondary structure of cell proteins by the presence of
epibrassinolide.
However no significant effect of epibrassinolide on cultured prostate epithelial
cells was observed.
Multivariate statistical analyses clearly indicated the differences between
epibrassinolide containing prostate cell lines and their control groups.
FTIR AND EDXRF INVESTIGATIONS
OF THE SECOND GENERATIONS OF
SALT TOLERANT SOYBEAN
MUTANTS
Sevim Akyuz*, Tanil Akyuz*, Ozge Celik**,
and Cimen Atak**
*Physics Department, Science and Letters Faculty, Istanbul Kultur
University, Atakoy Campus, Bakirkoy 34156, Istanbul, Turkey.
**Molecular Biology and Genetics Department, Science and Letters
Faculty, Istanbul Kultur University, Atakoy Campus, Bakirkoy 34156,
Istanbul, Turkey.
In this study the elemental and molecular structural alterations of the
gamma induced mutants of salt tolerant soybean seeds were
investigated in comparison with those of control groups. The protein,
lipid and carbohydrate structure of the second (M2) generations of the
salt tolerant soybean seeds and control group were investigated by FTIR
spectrometry.
The seeds of S04-05 (Ustun 1)) variety divided into two groups. Firstly,
a group of the seeds were gamma irradiated with 150 Gy dose, using
Cs-137 gamma source in IBL 437C irradiation property at dose rate of
6.248 Gy/min while the other group of the seeds (control group) were
unirradiated. The irradiation doses have been selected according to
cause minimum physiological damages.
Salt tolerance studies were performed in M2 generation of irradiated seeds of S04-05 (Ustun 1) variety in vitro conditions.
FT-IR spectra of soybean seads; C = control group; M = Mutants
Lipid (3000-2800, ca.1700 cm-1); protein (1690-1530; 1300-1200 cm-1), cellulosic
compounds (ca.1450-1100 cm-1) and carbohydrate (ca.1200-900 cm-1) bands are
observed
From the original spectra of soybean seeds, it is impossible to find any differences between
the control group and mutants of salt tolerant soybean seeds
Band component analysis of CH stretching region of the IR spectrum of a control group
soybean seed. Gaussian function was used to quantify the multicomponent peak area.
3009 cm-1 band is characteristics CH stretching mode of starch
Soybean seed protein contains a mix structures , including α-helices, β-sheets and β-turns.
When the ratios of α-helices and β-sheets differ in the protein structure, protein nutritive
value may also differ [4].Since proteins specific susceptibility to inhibitory activities in the
gastrointestinal tract is highly associated with its secondary structure
Second derivatives of the FTIR difference spectra obtained by subtracting the IR
spectrum of water from those of soybean seads.
Band component analysis of the 1700-1500 cm-1 region of the IR spectrum of M
150-2 after subtracted water bands.
Band component analysis of the 1700-1500 cm-1 region of IR spectrum of control
group soybean seed after subtracted water bands.
1746 cm-1 band corresponds to the pectin ester C=O stretching
1652-1653 cm-1 ; amid1 alpha helix, 1540-1544 cm-1 ; amid II alpha helix
1175 cm-1 ; cellulose
1124-1114; 1019 cm-1 characteristic bands of pectic polysaccharides,
1072 cm-1 carbohydrates (C-C, C-O stretching)
724 cm-1 (CH2) mode of lipids
The Infrared difference spectra between salt tolarant
mutant and control soybean seeds indicate that lipid and
cellulose components increase but protein
(particularly α-helix) structure decreases in mutant seeds in
comparison to control seeds.
C
1480-880 cm-1 region
Protein alpha helices structure is strongly positively and lipid is negatively associated with PC1.
In order to reveal the dissimilarities between control
group seeds and salt tolarant mutants, Hierarchical
Average linkage cluster analysis were performed on
the 3030-2800 and 1800-900 cm-1 regions of the
vector normalized IR spectra (water bands were
subtracted) using Euclidean distance.
2 point straight baseline corrections between 30302800 cm-1, 1800-1482 cm-1, 1482-1202 cm-1, 1202-900
cm-1 regions, were applied.
The dendrongram is as follows:
Cluster analysis of molecular spectra (distance method; Euclidean; Cluster method;
Hierarchical Average linkage)
Conclusion
• The genetic improvement studies done on quality characters of
soybean are serious research issues when the global consumption
of this plant has been thought . In this study the protein, lipid and
carbohydrate structures of the second (M2) generations of the salt
tolerant soybean seeds were investigated in comparison with the
control by group FTIR spectrometry.
• Results indicated the presence of some alterations in the secondary
structure of proteins in M2 mutants comparison to those of control
groups. β sheet to α helix ratio is found to increase in going from
control seeds to mutants.
• Protein quality relies not only on total protein and amino acid
content but also on protein secondary structure. Changes in protein
secondary structure, namely the increase β sheet to α helix ratio
may affect the protein nutritive value.
• In order to reveal the data embedded in the molecular structure
alterations of salt tolarant soybean seeds in comparison with the
control group, we performed two chemometric methods, principle
component analysis (PCA) and hiararchical cluster analysis.
• Multivariate statistical analyses clearly indicated the differences
between mutants and control groups.
• EDXRF analysis of the samples indicated that salt tolarant mutant
seeds has more NaCl than control group seeds. Trace elements
concentrations of both control group seeds and mutant seeds are
almost the same except Zn content. Zn concentration is found to be
almost three times higher in mutant seeds.