week3
advertisement
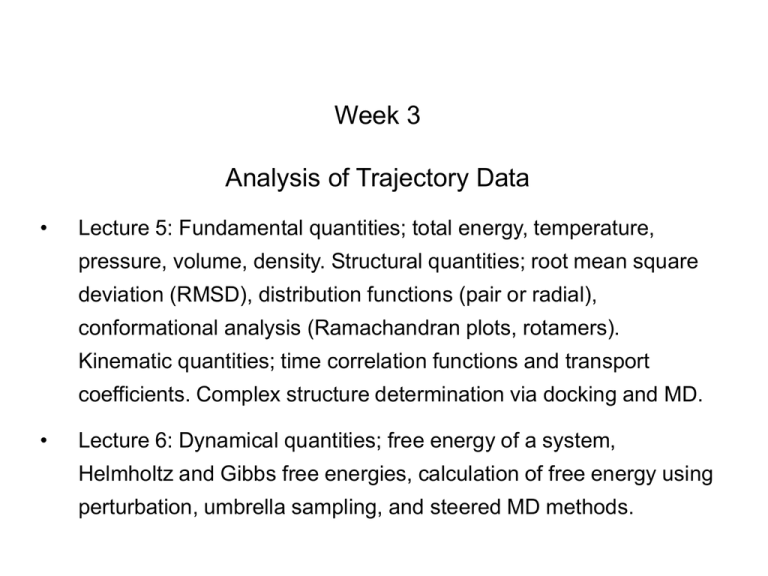
Week 3
Analysis of Trajectory Data
•
Lecture 5: Fundamental quantities; total energy, temperature,
pressure, volume, density. Structural quantities; root mean square
deviation (RMSD), distribution functions (pair or radial),
conformational analysis (Ramachandran plots, rotamers).
Kinematic quantities; time correlation functions and transport
coefficients. Complex structure determination via docking and MD.
•
Lecture 6: Dynamical quantities; free energy of a system,
Helmholtz and Gibbs free energies, calculation of free energy using
perturbation, umbrella sampling, and steered MD methods.
Structural analysis of proteins and complexes
1. Fundamental quantities
Total energy, temperature, pressure, volume (or density)
2. Structural quantities
Root Mean Square Deviation (RMSD)
Distribution functions (e.g., pair and radial)
Conformational analysis (e.g., Ramachandran plots, rotamers)
3. Complex structure determination
Determination of protein-ligand and protein-protein complexes using
docking programs.
Refinement of complex structures using MD simulations.
Fundamental quantities
• Total energy: strictly conserved but due to numerical errors it may drift.
• Temperature: would be constant in a macroscopic system (fluctuations
are proportional to 1/N, hence negligible). But in a small system,
they will fluctuate around a base line. Temperature coupling ensures
that the base line does not drift from the set temperature.
• Pressure: similar to temperature, though fluctuations are much larger
compared to the set value of 1 atm.
• Volume (or density): fixed in the NVT ensemble but varies in NPT.
Therefore, it needs to be monitored during the initial equilibration to
make sure that the system has converged to the correct volume
(density). Relatively quick for globular proteins in water but could take
a long time (~1 ns) for systems involving lipids (membrane proteins).
Root Mean Square Deviation
For an N-atom system, variance and RMSD at time t are defined as
1 N
(t ) ri (t ) ri (0)2 ,
N i 1
RMSD
Where ri(0) are the reference coordinates. Usually they are taken from the
first frame in an MD simulation, though they can also be taken from the
PDB or a target structure. Because side chains in proteins are different,
it is common to use a restricted set of atoms such as Ca or the backbone
(N, Ca, C,O) atoms.
RMSD is very useful in monitoring the approach to equilibrium (typically,
signalled by the appearance of a broad shoulder).
It is a good practice to keep monitoring RMSD during production runs
to ensure that the system stays near equilibrium.
Evolution of the RMSD for the backbone atoms of ubiquitin
Distribution functions
Pair distribution function gij(r) gives the probability of finding a pair of
atoms (i, j) at a distance r. It is obtained by sampling the distance rij in
MD simulations and placing each distance in an appropriate bin, [r, r+r].
Pair distribution functions are used in characterizing correlations between
pair of atoms, e.g., hydrogen bonds, cation-carbonyl oxygen. The peak
gives the average distance and the width, the strength of the interaction.
When the distribution is isotropic (e.g., ion-water in an electrolyte), one
samples all the atoms in the spherical shell, [r, r+r]. Thus to obtain the
radial distribution function (RDF), the sampled number needs to be
normalized by the volume ~4r2r (as well as density)
gion water (r )
N (r )
4r 2 r
Radial distribution functions exhibit oscillations, where each maxima is
identified with a coordination number. They are obtained by integrating
g(r) between two neighbouring minima, e.g. denoting the first minimum by
rmin1, the first coordination number is given by
N1
rmin1
2
4
r
g (r )dr
0
Similarly, the second coordination number can be found from
N2
rmin 2
2
4
r
g (r )dr
rmin1
where rmin2 denotes the second minimum in the RDF.
RDF for a Lennard-Jones liquid
Example: Binding of charybdotoxin to KcsA potassium channel
Important charge int’s:
K27 – Y78 (ABCD)
K11
R34
R34 – D80 (D)
R25 – D64, D80 (C)
K11 – D64 (B)
K27
Pair distribution functions for charge interactions
10
RMSD of charybdotoxin (ChTX)
11
Configurational changes in ChTx
during umbrella sampling simul.
The N-terminal moves away
and the helix is distorted
(L16-V20 H-bond is broken)
Changes are induced by the
tidal forces arising from the
competition between the
applied harmonic forces pulling
the toxin out and the Coulomb
forces pulling it in. Such
simulation artefacts has to be
avoided using restraints.
Conformational analysis
In proteins, the bond lengths and bond angles are more or less fixed.
Thus we are mainly interested in conformations of the torsional angles.
As the shape of a protein is determined by the backbone atoms, the
torsional angles, f (N−Ca) and y (C−Ca), are of particular interest.
These are conveniently analyzed using the Ramachandran plots.
Conformational changes in a protein during MD simulations can be most
easily revealed by plotting these torsional angles as a function of time.
Also of interest are the torsional angles of the side chains, c1, c2, etc.
Each side chain has several energy minima (called rotamers), which are
separated by low-energy barriers (~ kT). Thus transitions between
different rotamers is readily achievable, and they could play functional
roles, especially for charged side chains.
(Sasisekharan)
y
a-helix
f ~ 57o
y ~ 47o
f
Bound atoms or groups of atoms fluctuate around a mean value.
Most of the high-frequency fluctuations (e.g. H atoms), do not directly
contribute to the protein function. Nevertheless they serve as “lubricant”
that enables large scale motions in proteins (e.g. domain motions) that
do play a significant role in their function. As mentioned earlier, large
scale motions occur in a ~ ms to s time scale, hence are beyond the
present MD simulations. (Normal mode analysis provides an alternative.)
But torsional fluctuations occur in the ns time domain, and can be studied
in MD simulations using time correlation functions, e.g.
Cf (t )
f (0) f (t )
f (0)
2
These typically decay exponentially and such fluctuations can be
described using the Langevin equation.
Time correlation functions & transport coefficients
At room temperature, all the atoms in the simulation system are in a
constant motion characterized by their average kinetic energy: (3/2)kT.
Free atoms or molecules diffuse according to the relationship
r 2 (t ) 6 Dt
While this relationship can be used to determine the diffusion coefficient,
more robust results can be obtained using correlation functions, e.g.,
D is related to the velocity autocorrelation function as
1
D v (0) v (t ) dt
30
Similarly, conductance of charged particles is given by the Kubo formula
1
3VkT
J (0) J (t ) dt,
0
J qi vi
i
(current)
Complex structure determination
Few proteins perform their function alone. In most cases, they form
complexes with other proteins to become functional, or binding of a ligand
to a protein triggers its function.
• Hemoglobin (oxygen carrying protein) is a complex of four proteins
• Ligand-gated ion channels are opened by binding of a ligand
• Most drugs act by binding to a receptor protein, moderating its function
Structures of many biomolecules have been determined using x-ray
diffraction or nuclear magnetic resonance (NMR). However, only in a
handful of cases, structures of complexes have been determined
experimentally. Accurate determination of complex structures is essential
for understanding the function of proteins, and will have many
applications in pharmacology (rational drug design) and biotechnology.
Basic methods for complex structure determination
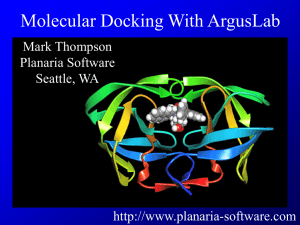
1. Docking and scoring: The simplest and the fastest method. Typically
an analytical function is used to estimate the interaction energy of the
protein-ligand complex for various positions, orientations and
conformations of the ligand, and its minimum value is used to find the
binding pose of the ligand. Limited accuracy, especially in scoring.
2. Brownian dynamics (BD) simulations: Trajectory of the ligand is
followed in BD simulations (implicit water) until it binds to the protein.
Slower than docking and provides no apparent advantages.
3. MD simulations: Trajectory of the ligand is followed in MD simulations
until it binds to the protein. Accurate but too slow to be useful in practice.
4. Docking combined with MD: Poses predicted by docking are refined in
MD simulations. Currently the most optimal method for this purpose.
Docking methods
AutoDock: The most popular docking method. Calculates the proteinligand interaction energies over a grid.
Scoring function: electrostatics + Lennard-Jones + hydrogen bond +
solvation + entropic term.
Protein is taken as rigid while ligand is allowed torsional flexibility.
Works well for small molecules (e.g. not good for > 30 AA peptide)
ZDOCK: Treats both protein and ligand as rigid, and hence can handle
much larger ligands. Searches for the binding position by optimizing
shape complementarity, electrostatics, and dehydration energies using a
Fast Fourier Transform (FFT) algorithm on a grid.
HADDOCK: Retains ligand flexibility and works for larger peptides. Much
more sophisticated (e.g. uses of exp. restraints, allows ensemble
docking, etc) but also much slower than the others.
Dynamical quantities – Free energy calculations
• Free energy of a simulation system
• Free energy difference between two states
• Calculation of free energy differences using free energy perturbation
(FEP) and thermodynamic integration (TI) methods; alchemical
transformation between two states
• Path dependent methods; potential of mean force along a reaction
coordinate; umbrella sampling with weighted histogram analysis
method (WHAM); steered MD with Jarzysnki’s equation.
• Calculation of binding free energies using path independent and path
dependent methods.
Free energy of a system
Free energy is the most important quantity that characterizes a dynamical
process. One can use either the Helmholtz or Gibbs free energy for this
purpose, which are given by
A = U – TS
(Helmholtz)
G = H – TS = U + pV – TS
(Gibbs)
In a typical biomolecular process, the volume and pressure do not
change, hence it is appropriate to use the Helmholtz free energy A.
Unfortunately, G has been used instead of A in the literature although the
calculated quantity is the Helmholtz free energy.
Calculation of the absolute free energies is difficult in MD simulations.
However, free energy differences can be estimated more easily and
several methods have been developed for this purpose.
Free energy perturbation (FEP)
The starting point for most approaches is Zwanzig’s perturbation formula for
the free energy difference between two states A and B, which are described
by the Hamiltonians HA and HB . Assuming that A and B differ by a small
perturbation, the free energy difference is given by:
G ( A B ) GB G A kT ln exp ( H B H A ) / kT
G ( B A) G A GB kT ln exp ( H A H B ) / kT
A
B
G ( A B ) G ( B A)
The equality should hold if there is sufficient sampling.
However, if the two states are not similar enough, this is difficult to achieve
in finite simulations, and there will be a large hysteresis effect.
(i.e. the forward and backward results will be very different)
Derivation of the perturbation formula
From statistical mechanics, the Helmholtz free energy is given by
G kT ln Z ,
Z
1
h
3N
N!
H / kT
e
dqdp
(Z: partition function)
Free energy difference between two states A and B can be written as
G ( A B) GB G A kT ln
H B / kT
e
dqdp
ZB
kT ln H / kT
ZA
e A dqdp
Inserting e H A / kTeH A / kT in the in top integral and using the probability
e H A / kT
PA (q, p) H / kT
A
e
dqdp
G ( A B) kT ln exp ( H B H A ) / kT PA (q, p ) dqdp
kT ln exp ( H B H A ) / kT
A
FEP with alchemical transformation
To obtain accurate results with the perturbation formula in finite time, the
energy difference between the states should be a few kT, which is not
satisfied for most biomolecular processes. To deal with this problem, one
introduces a hybrid Hamiltonian
H (l ) (1 l ) H A lH B
and performs the transformation from A to B gradually by changing the
parameter l from 0 to 1 in small steps. That is, one divides [0,1] into n
subintervals with {li, i = 0, n}, and for each li value, calculates the free
energy difference from the ensemble average
G(li li 1 ) kT ln exp[( H (li 1 ) H (li ))]/ kT
li
24
The total free energy change is then obtained by summing the contributions
from each subinterval
n 1
G (0 1) G (li li 1 )
i 0
The number of subintervals is chosen such that the free energy change at
each step is < a few kT, otherwise the method may lose its validity. Points
to be aware of:
1. Most codes use equal subintervals for li. But the changes in Gi
are usually highly non-linear. One should try to choose li such that
Gi remains around a few kT for all values, e.g., for charge
interactions, using exponential spacing gives better results.
2. The simulation times (equilibration + production) have to be chosen
carefully. It is not possible to extend them in case of non-convergence
25
(have to start over).
Thermodynamic integration (TI)
Another way to obtain the free energy difference is to integrate the
derivative of the hybrid Hamiltonian H(l):
H H / kT
e
dqdp
dG
H
l H / kT
dl
l
dqdp
e
1
l
G
0
H (l )
l
l
dl
This integral is evaluated most efficiently using a Gaussian quadrature.
In typical calculations for ions, 7-point quadrature is found to be sufficient.
(But one should check that other quadratures give the same result)
The advantage of TI over FEP is that the production run can be extended
as long as necessary and the convergence of the free energy can be
monitored (when the cumulative G flattens, it has converged).
26
Example : Binding of a K+ ion to a protein
Initial state (A): K+ ion in bulk, a water molecule at the binding site.
Final state (B): K+ ion at the binding site, water in place of the ion.
The free energy difference between A and B gives free energy gain in
translocating a K+ ion from bulk to the binding site of the protein. Note that
this is not the binding free energy – that includes entropic changes as well.
K+
W
protein
A
+ K+ (bulk)
protein
B
G( A B) G( B) G( A)
+ W (bulk)
Example: Free energy change due to mutation of a ligand
A very common question is how a mutation in a ligand (or protein) changes
the free energy of the protein-ligand complex.
GB G A Gbs ( A B ) Gbulk ( A B )
+
+
GA
Gbulk(AB)
+
GB
Thermodynamic cycle
wild type
Gbs(AB)
mutant
Standard binding free energy from FEP/TI methods
The total binding free energy of a ligand can be expressed as
Gb Gtr Grot Gres Gint
(2e) 3 / 2 x y z
Gtr kT ln
, V0 1660 3
V0
Grot
(2e) 3 / 2 f1 f 2 f 3
kT ln
2
8
bulk
site
Gres Gres
Gres
The various sigma’s are the translational and rotational rmsd’s of the ligand
in the binding site. The last term is the interaction (or translocation) energy,
which is calculated using FEP or TI.
29
Path dependent methods
Methods such as FEP and TI are independent of the physical path
connecting the two states. As seen in the previous example, calculation
of free energy difference for binding of a ligand involves annihilation of
the ligand in bulk and its creation in the binding site. Such calculations
can be done fairly accurately for small, uncharged ligands, but FEP/TI
methods fail for large or charged ligands. The reason is that the hydration
energies of such ligands are quite large and the errors committed in each
(bulk and binding site) calculation is usually larger than the free energy
difference between the two states.
To calculate the binding free energy of such ligands, one has to use path
dependent methods, where the ligand is physically moved from the
binding site to bulk along a reaction coordinate, and its free energy profile
along this path is calculated using various methods.
Potential of mean force (PMF)
Interaction of two molecules in solution is described by their PMF, W(r),
whose gradient gives the average force acting between them.
The PMF can be determined from MD simulations by sampling the relative
positions of the two molecules. Assuming one is at the origin, the density of
the second will be given by (r). From Boltzmann equation, the density is
given by
(r) (r0 ) e[W (r )W (r0 )] / kT
Here r0 is a reference point in bulk where W is assumed to vanish.
Inverting the Boltzmann equation yields
(r )
W (r ) W (r0 ) kT ln
(r0 )
Thus by sampling the density along a reaction coordinate, one can
determine the PMF between two molecules.
Umbrella sampling simulations
In general, position of a molecule cannot be adequately sampled at
high-energy points in finite simulations. To counter that, one introduces
harmonic potentials, which restrain the particle at desired points, and then
unbias its effect (this can be done easily for a harmonic biasing potential).
For convenience, one introduces umbrella potential at regular intervals
along a reaction coordinate (e.g. ~0.5 Å). After adequate sampling of each
window, the raw density data are unbiased, and the PMFs obtained from
all the windows are optimally combined using the Weighted Histogram
Analysis Method (WHAM).
For a simple introduction to the WHAM method and how it is implemented
in practice, see Alan Grossfield’s web page (search for “wham method” in
google and download the pdf file).
Points to consider in umbrella sampling
Two main parameters in umbrella sampling are the force constant, k
and the distance between windows, d. In bulk, the position of the
ligand will have a Gaussian distribution given by
1
( z z0 ) 2
P( z )
e
2
2 2
,
z z 0 , k BT / k
The overlap between two Gaussian distributions separated by d
% overlap 1 erf (d / 8 )
The parameters should be chosen such that the percentage overlap
satisfies, 10-20% > % overlap > 5%
If the overlap is too small, PMF will have discontinuities.
If it is too large, simulations are not very efficient.
Standard binding free energy from PMF
Experimentally, the standard binding free energy is determined by
measuring the dissociation constant Kd (or IC50), which is defined as the
ligand concentration at which half the ligands are bound. The standard
binding free energy is related to the binding constant Keq = 1/Kd via
Gb kT lnC0 K d kT lnC0 K eq
Where C0 is the standard concentration of 1 M (i.e., 1/ C0 = 1660 Å3)
The binding constant is determined by integrating the 3-D PMF over the
binding site
K eq
W (r )
e
kT 3
d r
site
Because it virtually impossible to determine the PMF in a 3-D grid, it is
34
necessary to invoke a 1-D approximation to the PMF.
1-D approximation of PMF
In most cases, there is a well-defined path from the binding site to bulk,
along which the PMF is determined. Thus the simplest approximation is to
perform the integral along this path assuming a flat-bottomed potential in the
transverse directions which will make a constant contribution. Taking the
path along the z-axis and assuming a cross sectional area of R2 for the
flat-bottomed potential, we obtain
K eq R 2
bulk
W ( z )
e
kT
dz
bs
The most sensible choice for R is to determine it from the transverse
fluctuations of the COM of the ligand in the binding pocket. In fact, this
choice can be rigorously justified using the simulation data without assuming
a flat-bottomed potential.
35
Justification of the 1-D approximation of PMF
Distribution of the x and y components of the peptide COM follows a
Gaussian distribution in all of the PMF windows (Kv1.3-HsTx1 complex)
Justification of the 1-D approximation of PMF
Gaussian distribution in x-y directions:
( x, y ) 0 e
( x 2 y 2 ) / 2 2
Comparing with the Boltzmann distribution due to W(x,y)
( x, y) 0 eW ( x, y ) / kBT
shows that the transverse PMF can be represented by a harmonic pot.
1
k T
W ( x, y ) k ( x 2 y 2 ) with k B2
2
Assuming W(x,y,z) = W(x,y) + W(z), the x-y integrals separate from z
e
W ( x , y ) k BT
dxdy e
k ( x 2 y 2 ) 2 k BT
1-D formula for Keq follows upon using
dxdy 2
k BT
2 2
k
R 2 x2 y2 2 2
37
Justification of the 1-D approximation of PMF
For the validity of the 1-D
approximation, the average
transverse position of the ligand
and its fluctuations should not
deviate much from the binding
site.
B) The average transverse
position of the ligand in each
umbrella window.
C) The R values (fluctuations)
in each umbrella window.
38
Steered MD (SMD) simulations and Jarzynski’s equation
Steered MD is a more recent method where a harmonic force is applied
to the COM r of a peptide and the reference point r0 of this force is
pulled with a constant velocity v. It has been used to study binding of
ligands and unfolding of proteins. The discovery of Jarzynski’s equation
in 1997, enabled determination of PMF from SMD, which boosted its
applications.
f
Work done by
the harmonic force
Jarzynski’s equation:
W F.ds , F k[r (r0 vt )]
i
e G / kT e W / kT
This method seems to work well in simple systems and when G is large
but beware of its applications in complex systems!