Clustering II
advertisement

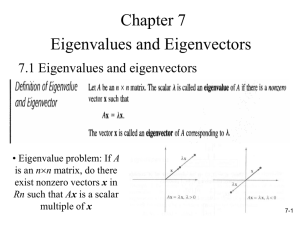
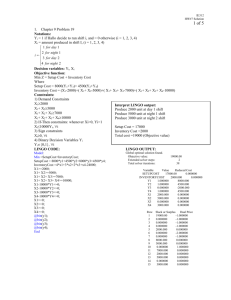
Clustering II CMPUT 466/551 Nilanjan Ray Mean-shift Clustering • Will show slides from: http://www.wisdom.weizmann.ac.il/~deniss/vision_spring04/files/mean_shift/mean_shift.ppt Spectral Clustering • Let’s visit a serious issue with K-means • K-means tries to figure out compact, hyperellipsoid like structures • What if the clusters are not ellipsoid like compact? K-means fails. 150 100 50 0 -50 -100 -150 -150 -100 -50 0 50 100 150 • What can we do? Spectral clustering can be a remedy here. Basic Spectral Clustering • Forms a similarity matrix wij for all pairs of observations i, j. • This is a dense graph with data points as the vertex set. Edge strength is given by the wij , similarity between ith and jth observations. • Clustering can be conceived as a partitioning of the graph into connected components, where within a component, the edge weights are large, whereas, across the components they are low. Basic Spectral Clustering… • Form the Laplacian of this graph: L G W , where G is a diagonal matrix N with entries, gi j 1Wij • L is positive semi-definite and has a constant eigenvector (all 1’s) with zero eigenvalue. • Find m smallest eigenvectors Z=[z1 z2 zm] of L, ignoring the constant eigenvector. • Cluster (say by K-means) N observations with features as rows of matrix Z. Why Spectral Clustering Works Insight 1: N N N 1 N N The graph cut cost for a label vector f: f Lf gi f i f i f j wij wij ( f i f j ) 2 2 i 1 j 1 i 1 i 1 j 1 T So, a small value of f T Lf will be obtained if pairs of points with large adjacencies same labels. Insight 2: The constant eigenvector corresponding to 0 eigenvalue is actually a trivial solution that suggests to put all N observations into a single cluster. If a graph has K connected components, the nodes of the graph can be reordered so that L will be block diagonal with K diagonal blocks and L will have zero eigenvalue with multiplicity K, one for each connected component. Corresponding eigenvectors will have indicator variables indentifying these connected components. In reality, we only have weak and strong edges. So look for small eigenvalues. Combining Insight 1 and 2: Choose eigenvectors corresponding to small eigenvalues and cluster them into K classes. A Tiny Example: A Perfect World W =[1.0000 0.5000 0 0 0 0 0.5000 0 0 1.0000 0 0 1.0000 0.8000 0.8000 1.0000]; L = 0.5000 -0.5000 0 0 0 0 -0.5000 0 0 0.5000 0 0 0.8000 -0.8000 -0.8000 0.8000]; We observe two classes each with 2 observations here. W is a perfect block diagonal matrix here. Laplacian L Eigenvalues of L: 0, 0, 1, 1.6 Eigenvectors corresponding to two 0 eigenvalues: [-0.7071 -0.7071 0 0] and [ 0 0 -0.7071 -0.7071] The Real World Tiny Example W =[ 1.0000 0.5000 0.0500 0.1000 0.5000 1.0000 0.0800 0.0400 0.0500 0.0800 1.0000 0.8000 0.1000 0.0400 0.8000 1.0000] L =[ 0.6500 -0.5000 -0.0500 -0.1000 -0.5000 0.6200 -0.0800 -0.0400 -0.0500 -0.0800 0.9300 -0.8000 -0.1000 -0.0400 -0.8000 0.9400] [V,D]=eig(L) Eigenvectors: Eigenvalues: V = 0.5000 0.5000 0.5000 0.5000 0.4827 0.5170 -0.5027 -0.4970 D = 0.0000 0.2695 -0.7169 0.6930 0.0648 -0.0409 0.0557 -0.0498 0.7022 -0.7081 1.1321 1.7384 Notice that eigenvalue 0 has a constant eigenvector. The next eigenvalue 0.26095 has an eigenvector that clearly indicates the class memberships. Normalized Graph Cut for Image Segmentation 5 10 15 20 25 30 35 40 45 50 10 20 30 40 50 60 A cell image Similarity: | I (i ) I ( j ) | 2 | X (i ) X ( j ) | 2 exp( ) exp( ), for | X (i ) X ( j ) | 25, W (i, j ) 2σ I2 2σ X2 0, otherwise. Pixel locations NGC Example 5 5 5 10 10 10 15 15 15 20 20 20 25 25 25 30 30 30 35 35 35 40 40 40 45 45 45 50 50 10 20 30 40 (a) 50 60 50 10 20 30 (b) 40 5 5 10 10 15 15 20 20 25 25 30 30 35 35 40 40 45 45 50 50 60 10 20 30 (c) 40 50 60 50 10 20 30 (d) 40 50 60 10 20 30 (e) 40 50 60 (a) A blood cell image. (b) Eigenvector corresponding to second smallest eigenvalue. (c) Binary labeling via Otsu’s method. (d) Eigenvector corresponding to third smallest eigenvalue. (e) Ternary labeling via k-means clustering. Demo: NCUT.m