PPT
advertisement

Power Iteration Clustering
Speaker: Xiaofei Di
2010.10.11
Outline
•
•
•
•
•
Authors
Abstract
Background
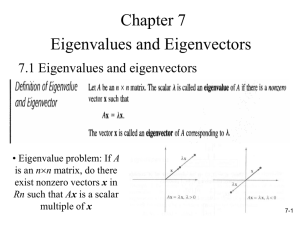
Power Iteration Clustering(PIC)
Conclusion
Authors
• Frank Lin
• PhD Student
Language Technologies Institute
School of Computer Science
Carnegie Mellon University
• http://www.cs.cmu.edu/~frank/
•
William W. Cohen
• Associate Research Professor, Machine
Learning Department, Carnegie Mellon
University
• http://www.cs.cmu.edu/~wcohen/
Abstract
• We present a simple and scalable graph clustering
method called power iteration clustering.
• PIC finds a very low-dimensional embedding of a
dataset using truncated power iteration on a
normalized pair-wise similarity matrix of the data. This
embedding turns out to be an effective cluster
indicator, consistently outperforming widely used
spectral methods such as Ncut on real datasets.
• PIC is very fast on large datasets, running over 1000
times faster than an Ncut implementation based on the
state-of-the-art IRAM eigenvector computation
technique.
摘要
• 本文提出了一种简单可扩展的图聚类方法:
快速迭代聚类(PIC)。
• PIC利用数据归一化的逐对相似度矩阵,采
用截断的快速迭代法,寻找数据集的一个
超低维嵌入。这种嵌入恰好是很有效的聚
类指标,使它在真实的数据集上总是好于
广泛使用的谱聚类方法,比如NCut。
• 在大规模数据集上,PIC非常快,比基于最
好的特征计算技术实现的Ncut快1000倍。
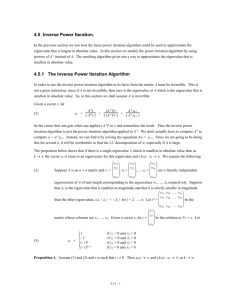
Background 1
----spectral clustering
dataset : X { x , x , ..., x }
1 2
n
similarity function : s ( x , x ) 0
i j
normalized affinity matrix: NA = D1W
unnormalized graph Laplacian matrix: L = D - W
affinity matrix : W s ( x , x )
ij
i j
normalized symmetric Laplacian matrix: L=I - D WD
deg ree matrix : D diagonal matrix
normalized random-walk Laplacian matrix: L=I - D-1W
d
ii
jW
ij
1
2
1
2
Background 2
----Power Iteration Method
• An eigenvalue algorithm
– Input: initial vector b0 and the matrix A
Ab
– Iteration: b Ab
k 1
k
k 1
•
Advantage
•
dose not compute matrix decomposition
Disadvantages
finds only the largest eigenvalue and converges slowly
•
Convergence
Under the assumptions:
A has an eigenvalue that is strictly greater in magnitude than its other
eigenvalues
The starting vector b0 has a nonzero component in the direction of an
eigenvector associated with the dominant eigenvalue.
then:
A subsequence of bk converges to an eigenvector associated with the
dominant eigenvalue
Power Iteration Clustering(PIC)
Unfortunately,
since the sum of each row of NA is 1, the largest eigenvector of NA (the smallest of L)
is a constant vector with eigenvalue 1.
Fortunately,
the intermediate vectors during the convergence process are interesting.
Example:
Conclusion:
PI first converges locally within a cluster.
xi R and s(x i ,x j )=exp(
2
|| xi x j ||2
2 2
)
PI’s Convergence
Let:
W = NA (Normalized affinity matrix ), vt is the t-th iteration of PI
W has eigenvectors e1 ,..., en with eigenvalues λ1 ,...,λ n
λ1 =1, e1 is constant
2 ,...., k are lager than the remaining ones
Spectral representation of
Spectral distance between a and b:
W has an ( , )-eigengap between the k th and (k+1)th eigenvector
every W is e bounded
(t, v0 ) distance between a and b:
k
2
and
k 1
2
j
[e j (a) e j (b)]c j
j k 1
2
n
t
signalt is an approximation of spec, but
a) compressed to the small radius R t
b) has components distorted by ci and i 2
t
c) has terms that are additively combined(rather than Euclidean)
a) The size of the radius is of no importance in clustering, because most clustering
methods based on relative distance, not absolute one.
b) The importance of the dimension associated with the i-th eigenvector is
downweighted by (a power of) its eigenvalue, which often improves performance
for spectral methods.
c) For many natural problems, W is approximately block-stochastic, and hence the
first k eigenvectors are approximately piecewise constant over the k clusters.
It is easy to see that when spec(a,b) is small, signal must also small. However, when
a and b are in different clusters, since the terms are signed and additively combined,
it is possible that they may “cancel out” and make a and b seem to be in the same
cluster. Fortunately, this seems to be uncommon in practice when the cluster
number k is not too large.
So for large enough a, small enough t,
signalt is very likely a good cluster indicator.
Early stopping for PI
velocity at t: t vt vt 1
acceleration at t: t t t 1
if || t || ˆ then stop PI
While the clusters are ‘’locally
converging”, the rate of convergence
changes rapidly; whereas during the
final global convergence, the
converge rate appears more stable.
1*105
1. ˆ
n
n is the number of data instances
2. v (i )
0
j
Aij
V ( A)
V(A)= i j Aij
3. V [v t1 ,..., v tk ] v t
(one dimension is good enough)
Experiments (1/3)
• Purity : cluster purity
• NMI : normalized mutual information
• RI : rand index
The Rand index or Rand measure is a measure of the similarity between two
data clusterings. Given a set of n elements
and two partitions
of S to compare,
and
, we define the following:
a = | S * | , where
b = | S * | , where
c = | S * | , where
d = | S * | , where
for some
Then:
Experiments (2/3)
Experimental comparisons on accuracy of PIC
Experimental comparisons on eigenvalue weighting
Experiments (3/3)
Experimental comparisons on scalability
Synthetic dataset
NCutE uses slower, classic eigenvalue decomposition method to find all eigenvectors.
NCutI uses fast Implicitly Restarted Arnoldi Method(IRAM) for the top k eigenvectors.
Conclusion
• Novel
• Simple
• Efficient
Appendix
----NCut
Appendix
----NJW