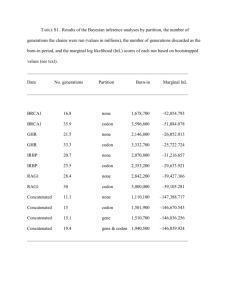
Ab_initio_predition_tools - Compgenomics2010
advertisement

GENE MARK
•Developed in 1993 at Georgia Institute of Technology as the first
gene finding tool.
•Used markov chain to represent the statistics of coding and
noncoding reading frames using dicodon statistics.
Shortcomings:
•Inability to find exact gene boundaries.
GENE MARK.hmm
• Improved gene prediction in terms of finding accurate gene boundaries.
• Incorporates use of Hidden Markov Model with duration..
• The aim of gene finding was to find the true functional sequence X from
the anonymous DNA sequence S.
S= {b1,b2,……………..,bL}
sequence.
X={x1,x2,……………..,xL}
complementary strand
bi stands for nucleotides A,G,C,T and L length of
xi
= 0 if nucleotide is in non coding region
= 1 if nucleotide is in coding region
= 2 if nucleotide is in coding region in
• Probability of any sequence S underlying functional sequence X is
calculated as P(X|S)=P(x1,x2,……………..,xL| b1,b2,……………..,bL)
• Viterbi algorithm then calculates the functional sequence X* such that
P(X*|S) is the largest among all possible values of X.
• Ribosome binding site model was also added to augment accuracy in the
prediction of translational start sites.
Even in prokaryotic genomes gene overlaps are quite common
RBS feature overcomes this problem by defining a % position nucleotide
matrix based on alignment of 325 E coli genes whose RBS signals have
already been annotated.
Uses a consensus sequence AGGAG to search upstream of any alternative
start codons for genes predicted by HMM.
GLIMMER
maintained by Steven Salzberg, Art Delcher at the University of
Maryland , College Park
•Used IMM (Interpolated markov Models) for the first time.
• Predictions based on variable context(oligomers of variable
lengths).
•More flexible than the fixed order Markov models.
•Three versions:
Glimmer 1 (1997)
Glimmer 2 (1999)
Glimmer 3 (2007)*
Principle:
IMM combines probability based on 0,1……..k
previous bases, in this case k=8 is used. But
this is for oligomers that occur frequently
however for rarely occurring oligomers 5 th
order or lower may also be used.
Glimmer 2
Glimmer 2 adds the concept of Interpolated context model to IMM.
ICM – More flexible and will choose any base in the variable context (not
only ones next to bk+1) to determine the probability of our base of interest
(bk+1) .
Based on codon bias in translation .
Add on Features:
- Increase in the sensitivity.
-
Glimmer 3:
• Overcomes the shortcomings of previous models by taking in account sum
of RBS score ,IMM coding potentials and a score for start codons which is
dependent on relative frequency of each possible start codon in the same
training set used for RBS determination.
• Algorithm used reverse scoring of IMM by scoring all ORF (open reading
frames) in reverse ,from the stop codon to start codon , with the
probability of each base conditioned on a context window on it’s 3’ side
and score being the sum of log likelihood of the bases contained in the
ORF
Highlights:
• Increase in sensitivity along with increase in specificity as well.
• Reduced overlapping predictions.
• Separating sequences from separate genomes.
• Better prediction for GC rich genomes .