Bacterial Gene Finding and Glimmer (also Archaeal and viral gene finding)
advertisement

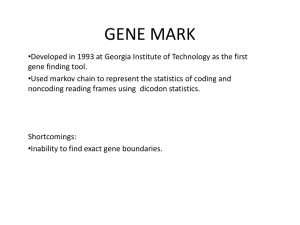
Bacterial Gene Finding and Glimmer (also Archaeal and viral gene finding) Arthur L. Delcher and Steven Salzberg Center for Bioinformatics and Computational Biology University of Maryland Outline • A (very) brief overview of microbial gene-finding – Codon composition methods – GeneMark: Markov models • Glimmer1 & 2 – Interpolated Markov Model (IMM) – Interpolated Context Model (ICM) • Glimmer3 – Reducing false positives – Improving coding initiation site predictions – Running Glimmer3 Step One • Find open reading frames (ORFs). …TAGAAAAATGGCTCTTTAGATAAATTTCATGAAAAATATTGA… Stop codon Stop codon Step One • Find open reading frames (ORFs). Reverse strand Stop codon …ATCTTTTTACCGAGAAATCTATTTAAAGTACTTTTTATAACT… …TAGAAAAATGGCTCTTTAGATAAATTTCATGAAAAATATTGA… Stop codon Shifted Stop • But ORFs generally overlap … Stop codon Campylobacter jejuni RM1221 30.3%GC All ORFs on both strands shown - color indicates reading frame Longest ORFs likely to be protein-coding genes Note the low GC content Campylobacter jejuni RM1221 30.3%GC Purple lines are the predicted genes Purple ORFs show annotated (“true”) genes Campylobacter jejuni RM1221 30.3%GC Mycobacterium smegmatis MC2 67.4%GC Note what happens in a high-GC genome Campylobacter jejuni RM1221 30.3%GC Mycobacterium smegmatis MC2 67.4%GC Purple lines show annotated genes The Problem • Need to decide which orfs are genes. – Then figure out the coding start sites • Can do homology searches but that won’t find novel genes – Besides, there are errors in the databases • Generally can assume that there are some known genes to use as training set. – Or just find the obvious ones Codon Composition • Find patterns of nucleotides in known coding regions (assumed to be available). – Nucleotide distribution at 3 codon positions – Hexamers – GC-skew • (G-C)/(G+C) computed in windows of size N – Amino-acid composition • Use these to decide which orfs are genes. – Prefer longer orfs – Must deal with overlaps Bacterial Replication Early replication Theta structure Termination of Replication E. coli B. subtilis Borrelia burgdorferi (Lyme disease pathogen) GC-skew plot Codon Composition Nucleotide variation at codon position: Mycobacterium smegmatis Campylobacter jejuni Codon Position Codon Position 1 2 a 19% 23% 6% 9% c 27% 28% 48% 14% 10% g 42% 20% 39% 33% 44% t 12% 28% 7% 1 2 3 a 36% 36% 36% c 13% 17% g 30% t 21% 3 Codon-Composition Gene Finders • ZCURVE – Guo, Ou & Zhang, NAR 31, 2003 – Based on nucleotide and di-nucleotide frequency in codons – Uses Z-transform and Fisher linear discriminant • MED – Ouyang, Zhu, Wang & She, JBCB 2(2) 2004 – Based on amino-acid frequencies – Uses nearest-neighbor classification on entropies Probabilistic Methods • Create models that have a probability of generating any given sequence. • Train the models using examples of the types of sequences to generate. • The “score” of an orf is the probability of the model generating it. – Can also use a negative model (i.e., a model of nonorfs) and make the score be the ratio of the probabilities (i.e., the odds) of the two models. – Use logs to avoid underflow Fixed-Order Markov Models • k th-order Markov model bases the probability of an event on the preceding k events. • Example: With a 3rd-order model the probability of this Target sequence: Context CTAGAT would be: P(G | CTA) P(A | TAG) P(T | AGA) Target Context Fixed-Order Markov Models • Advantages: – Easy to train. Count frequencies of (k+1)-mers in training data. – Easy to compute probability of sequence. • Disadvantages: – Many (k+1)-mers may be undersampled in training data. – Models data as fixed-length chunks. Fixed-Length Context Target …ACGTAGTTCAGTA… GeneMark • Borodovsky & McIninch, Comp. Chem 17, 1993. • Uses 5th-order Markov model. • Model is 3-periodic, i.e., a separate model for each nucleotide position in the codon. • DNA region gets 7 scores: 6 reading frames & non-coding―high score wins. • Lukashin & Borodovsky, Nucl. Acids Res. 26, 1998 is the HMM version. Interpolated Markov Models (IMM) • Introduced in Glimmer 1.0 Salzberg, Delcher, Kasif & White, NAR 26, 1998. • Probability of the target position depends on a variable number of previous positions (sometimes 2 bases, sometimes 3, 4, etc.) • How many is determined by the specific context. ggtta • E.g., for context ggtta the next position might depend on previous 3 bases tta . But for context catta all 5 bases might be used. Real IMMs • Model has additional probabilities, λ, that determine which parts of the context to use. • E.g., the probability of g occurring after context atca is: (atca)P (g | atca) (1 (atca))[(tca)P (g | tca) (1 (tca))[(ca)P (g | ca) (1 (ca))[ (a)P (g | a) (1 (a))P (g)]]] Real IMMs • Result is a linear combination of different Markov orders: b4P(g | atca) b3P(g | tca) b2P(g | ca) b1P(g | a) b0P(g) where b0 b1 b2 b3 b4 1 • Can view this as interpolating the results of different-order models. • The probability of a sequence is still the probability of the bases in the sequence. Real IMMs • Problem: How to determine the λ’s (or equivalently the bj’s)? • Traditionally done with EM algorithm using cross-validation (deleted estimation). – Slow – Hard to understand results – Overtraining can be a problem • We will cover EM later as part of HMMs Glimmer IMM • Glimmer assumes: – Longer context is always better – Only reason not to use it is undersampling in training data. • If sequence occurs frequently enough in training data, use it, i.e., 1 • Otherwise, use frequency and χ2 significance to set λ. • Interpolation is always between only 2 adjacent model lengths. More Precisely • Suppose context of length k+1 occurs a times, a<400, with target distribution D1; and context of length k occurs ≥400 times, with target distribution D2 • Let p be the χ2 probability that D1 differs from D2 • Then the interpolated distribution will be: pa pa D1 1 D2 400 400 This formula computes the probability of a base B at any position in a sequence, given the context preceding that position This formula is repeatedly invoked for all positions in an ORF IMMs vs Fixed-Order Models • Performance – IMM generally should do at least as well as a fixedorder model. – Some risk of overtraining. • IMM result can be stored and used like a fixedorder model. • IMM will be somewhat slower to train and will use more memory. Variable-Length Context Target …ACGTAGTTCAGTA… Interpolated Context Model (ICM) • Introduced in Glimmer 2.0 Delcher, Harmon, et al., Nucl. Acids Res. 27, 1999. • Doesn’t require adjacent bases in the window preceding the target position. • Choose set of positions that are most informative about the target position. Variable-Position Context Target …ACGTAGTTCAGTA… ICM • For all windows compare distribution at each context position with target position ************* Compare • Choose position with max mutual information p ( x, y ) I ( X ; Y ) p( x, y ) log p( x) p( y ) x y ICM • Continue for windows with fixed base at chosen A positions **** ******** Compare • Recurse until too few training windows – Result is a tree—depth is # of context positions used • Then same interpolation as IMM, between node and parent in tree Sample ICM Model ver = 2.00 len = 12 depth = 7 0 ---|---|---|*-? 0.0260 1 ---|---|---|a*? 0.0769 2 ---|---|---|c*? 0.0243 3 ---|---|---|g*? 0.0597 4 ---|---|---|t*? 0.0277 5 ---|---|--*|aa? 0.0051 6 ---|---|--*|ac? 0.0046 7 ---|---|--*|ag? 0.0101 8 ---|---|*--|at? 0.0037 9 ---|---|--*|ca? 0.0019 10 ---|---|--*|cc? 0.0090 11 ---|---|--*|cg? 0.0035 12 ---|---|--*|ct? 0.0083 13 ---|---|--*|ga? 0.0057 14 ---|---|--*|gc? 0.0057 15 ---|---|--*|gg? 0.0056 16 ---|---|--*|gt? 0.0096 17 ---|---|--*|ta? 0.0034 18 ---|---|--*|tc? 0.0062 19 ---|---|--*|tg? 0.0052 20 ---|---|-*-|tt? 0.0076 21 ---|---|-*a|aa? 0.0035 22 ---|---|-*c|aa? 0.0037 23 ---|---|-*g|aa? 0.0052 24 ---|--*|--t|aa? 0.0060 25 ---|---|-*a|ac? 0.0031 periodicity = 0.225 0.240 0.243 0.165 0.237 0.294 0.219 0.240 0.209 0.245 0.320 0.202 0.195 0.118 0.180 0.207 0.236 0.147 0.199 0.246 0.297 0.189 0.180 0.406 0.279 0.327 0.277 0.326 0.233 0.158 0.119 0.252 0.207 0.232 0.205 0.170 0.179 0.238 0.183 0.338 0.244 0.244 0.376 0.199 0.313 0.197 0.283 0.199 0.259 0.231 0.176 0.138 3 nodes = 21845 0.358 0.177 0.501 0.092 0.264 0.205 0.407 0.134 0.291 0.256 0.478 0.000 0.541 0.146 0.613 0.000 0.400 0.217 0.303 0.253 0.308 0.207 0.267 0.148 0.189 0.206 0.397 0.000 0.473 0.136 0.417 0.213 0.359 0.203 0.400 0.224 0.290 0.293 0.323 0.156 0.194 0.318 0.425 0.000 0.490 0.000 0.517 0.000 0.510 0.000 0.519 0.166 ICM • ICM’s not just for modeling genes • Can use to model any set of training sequences – Doesn’t even need to be DNA • Have used recently to finding contaminant sequences in shotgun sequencing projects. – Used a non-periodic (i.e., homogeneous) model Fixed-Length Sequences and ICMs • Array of ICM’s―a different one for each position in fixed-length sequence. • Can model fixed regions around transcription start sites or splice sites. • Positions in region can be permuted. ACGTAGTTCAGTAG ACGTAGTTCAGTAG Overlapping Orfs • Glimmer1 & 2 used rules. • For overlapping orfs A and B, the overlap region AB is scored separately: – If AB scores higher in A’s reading frame, and A is longer than B, then reject B. – If AB scores higher in B’s reading frame, and B is longer than A, then reject A. – Otherwise, output both A and B with a “suspicious” tag. • Also try to move start site to eliminate overlaps. • Leads to high false-positive rate, especially in high-GC genomes. Glimmer 2.0 Overlap Comments OrfID 2 3 4 5 7 8 9 10 11 12 13 15 16 17 18 19 20 21 23 25 26 27 28 29 30 31 32 33 Start 499 1977 1721 2624 3775 4501 6633 6648 9229 10411 11215 12827 12243 13298 15143 15193 15519 17249 19052 42693 41623 42899 43351 45223 45261 46261 46701 46752 End 1689 418 2611 3775 4356 6615 4513 9173 10008 11208 12243 11379 13298 14137 14133 15519 17249 19015 41620 41692 43065 43351 44685 45747 44695 45857 46420 47543 Frame Len Score [+1 L=1191 r=-1.151] [-1 L=1560 r=-1.317] [+2 L= 891 r=-1.156] [+2 L=1152 r=-1.184] [+1 L= 582 r=-1.223] [+1 L=2115 r=-1.181] [-1 L=2121 r=-1.365] [+3 L=2526 r=-1.155] [+1 L= 780 r=-1.217] [+1 L= 798 r=-1.192] [+1 L=1029 r=-1.165] [-3 L=1449 r=-1.295] [+3 L=1056 r=-1.228] [+2 L= 840 r=-1.161] [-3 L=1011 r=-1.230] [+1 L= 327 r=-1.284] [+3 L=1731 r=-1.175] [+2 L=1767 r=-1.266] [+2 L=22569 r=-1.144] [-1 L=1002 r=-1.200] [+1 L=1443 r=-1.401] [+2 L= 453 r=-1.261] [+1 L=1335 r=-1.169] [+1 L= 525 r=-1.153] [-1 L= 567 r=-1.298] [-2 L= 405 r=-1.275] [-1 L= 282 r=-1.273] [+3 L= 792 r=-1.178] Comments [ShadowedBy #3] [Contains #2] [LowScoreBy #4 L=257 S=0] [OlapWith #3 L=257 S=99] [DelayedBy #5 L=216] [OlapWith #9 L=2103 S=99] [LowScoreBy #8 L=2103 S=0] [ShorterThan #15 L=865 S=99] [LowScoreBy #13 L=865 S=0] [LowScoreBy #16 [OlapWith #15 L=585 S=99] [DelayedBy #13 L= [DelayedBy #20 L=1263] [ShadowedBy #26] [Contains #25] [LowScoreBy #27 L=167 S=0] [ShorterThan #26 L=167 S=99] [DelayedBy #29 L=363] Glimmer3 • Uses a dynamic programming algorithm to choose the highest-scoring set of orfs and start sites. • Not quite an HMM – Allows small overlaps between genes • “small” is user-defined – Scores of genes are not necessarily probabilities. – Score includes component for likelihood of start site Reverse Scoring • In Glimmer3 orfs are scored from 3’ end to 5’ end, i.e., from stop codon back toward start codon. • Helps find the start site. – The start should appear near the peak of the cumulative score in this direction. – Keeps the context part of the model entirely in the coding portion of gene, which it was trained on. Reverse Scoring Finding Start Sites • Can specify a position weight matrix (PWM) for the ribosome binding site. • Used to boost the score of potential start sites that have a match. • Would like to try: – A fixed-length ICM model of start sites. – A decision-tree model of start sites. – Hard to get enough reliable data. • Glimmer2 had a hybridization-energy calculation with 16sRNA tail sequence that didn’t work well. Glimmer3 vs. Glimmer2 Table 3. Probability models trained on the output of the long-orfs program. Predictions compared to genes with annotated function. Table 4. Probability models trained on the output of the long-orfs program. Predictions compared to all annotated genes. Other Glimmer3 Features • Can specify any set of start or stop codons – Including by NCBI translation table # • Can specify list of “guaranteed” genes – Or just orfs and let Glimmer choose the starts • Can specify genome regions to avoid – Guarantee no prediction will overlap these regions – Useful for RNA genes (tRNAs, rRNAs) • Output more meaningful orf scores. • Allow genes to extend beyond end of sequence – important for annotation of incomplete genomes Glimmer3 Output ID 0007 0008 0009 0010 0011 0012 0013 Frame ... -2 -3 -3 -2 -3 +3 -2 +1 -2 -3 -2 -2 +3 +1 -3 +1 +2 -2 +3 -2 +1 +1 -3 +3 +1 -2 -2 -2 ----- Start ----of Orf of Gene ... ... 10516 10459 10787 10781 11132 11120 12058 12055 12437 12371 8109 8142 12763 12724 12850 12877 13165 13036 13649 13634 13675 13663 13972 13972 12633 12642 14194 14323 14669 14663 14482 14548 14570 14642 14947 14935 14655 14718 15115 15100 15004 15070 15364 15475 15701 15671 15531 15591 15631 15649 15922 15898 16561 16561 16741 16711 Stop ... 10337 10629 11004 11936 12249 12632 12545 12996 12905 13473 13256 13850 14087 14454 14088 14673 14806 14696 14954 14975 15288 15594 15000 15728 15762 15719 16307 16562 --- Length ---of Orf of Gene ... ... 177 120 156 150 126 114 120 117 186 120 4521 4488 216 177 144 117 258 129 174 159 417 405 120 120 1452 1443 258 129 579 573 189 123 234 162 249 237 297 234 138 123 282 216 228 117 699 669 195 135 129 111 201 177 252 252 177 147 -------------- Scores -----------Raw Frame +1 +2 +3 -1 -2 -3 NC ... ... 1.14 0 - - 99 - 0 - 0 -10.69 0 - - 99 - - 0 0 -13.54 0 - - 99 - - 0 0 -2.25 0 - - 99 - 0 - 0 -13.68 0 - - 99 0 - 0 0 16.05 99 - - 99 - - - 0 4.17 99 - - - - 99 - 0 -16.13 0 0 - 99 - - - 0 -1.31 0 - - 99 - 0 - 0 -7.11 0 - - 99 - 0 0 0 2.83 0 - - 99 - 0 - 0 4.25 0 - - 99 - 0 - 0 15.20 99 - - 99 - - - 0 1.62 0 0 - - - - 99 0 13.36 99 - - - - - 99 0 3.07 0 0 - - - - 99 0 -4.35 0 - 0 - - - - 99 16.17 99 - - 0 - 99 - 0 3.06 99 - - 99 - - - 0 -7.02 0 - - - - 0 - 99 -2.46 0 0 - - - - 99 0 -5.48 0 0 - - - - 99 0 13.07 99 - - - - - 99 0 -8.84 0 - - 0 - - - 99 -0.69 3 3 - - - - - 96 -0.41 6 - - - - 6 - 93 1.73 0 - - 99 - 0 - 0 1.72 0 - - 99 - 0 - 0 Finding Initial Training Set • Glimmer1 & 2 have program long-orfs • It finds orfs longer than min-length that do not overlap other such orfs. – Current version automatically finds min-length. • Works OK for low- to medium-GC genomes • Terrible for high-GC genomes – Up to half the orfs can be wrong. – Uses first possible start site―even if orf is correct much of sequence isn’t. • Can use codon composition information in Glimmer3 version (similar to MED). Running Glimmer3 #1 Find long, non-overlapping orfs to use as a training set long-orfs --no_header -t 1.0 tpall.1con tp.longorfs #2 Extract the training sequences from the genome file extract --nostop tpall.1con tp.longorfs > tp.train #3 Build the icm from the training sequences build-icm -r tp.icm < tp.train #4 Run first Glimmer glimmer3 -o50 -g110 -t30 tpall.1con tp.icm tp.run1 #5 Get training coordinates from first predictions tail +2 tp.run1.predict > tp.coords #6 Create a position weight matrix (PWM) from the regions # upstream of the start locations in tp.coords upstream-coords.awk 25 0 tp.coords | extract tpall.1con - > tp.upstream elph tp.upstream LEN=6 | get-motif-counts.awk > tp.motif #7 Determine the distribution of start-codon usage in tp.coords set startuse = `start-codon-distrib --3comma tpall.1con tp.coords` #8 Run second Glimmer glimmer3 -o50 -g110 -t30 -b tp.motif -P $startuse tpall.1con tp.icm tp A novel application of Glimmer • P. didemni is a photosynthetic microbe that lives as an endosymbiont in the sea squirt > patella • P. didemni can only be cultured in L. patella cells L. patella (sea squirt) A novel application of Glimmer • • • • Generated 82,337 shotgun reads Bacterial genome 5 Mb Host genome estimated at 160 Mb Depth of coverage therefore much greater for bacterial contigs • Singleton reads primarily belong to host A novel application of Glimmer • Create training sets by classifying reads from scaffolds > 10kb as bacterial – 36,920 reads • Reads where both read and mate were singletons were treated as sea squirt – 21,276 reads • 21,141 reads unclassified A novel application of Glimmer • Train a non-periodic IMM on both sets of data • 2 IMMs created • Then classify reads using the ratio of scores from the two IMMs • In a 5-fold cross-validation, classification accuracy was – 98.9% on P. didemni reads – 99.9% on L. patella reads A novel application of Glimmer • Finally, re-assemble using ONLY bacterial reads • Original assembly: – 65 scaffolds of 20 Kb or longer – total scaffold length 5.74 Mb • Improved assembly: – 58 scaffolds of 20 Kb or longer – total scaffold length 5.84 Mb Acknowledgements Art Delcher Steven Salzberg Owen White (TIGR) Simon Kasif (Boston U.) Doug Harmon (Loyola College) Kirsten Bratke Edwin Powers (Johns Hopkins U.) Dan Haft (TIGR) Bill Nelson (TIGR)