Reconstruction and analysis of human liver-specific
advertisement
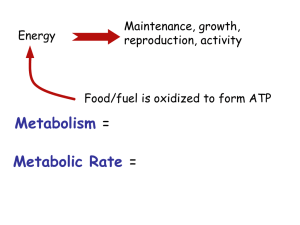
Reconstruction and analysis of human liver-specific metabolic network based on CNHLPP data Jing Zhao Logistical Engineering University The 6th Chinese Conference of Complex Networks,October 15-18, 2010. Suzhou, China OutLine • Background • Reconstruction of metabolic networks • Basic topological features of metabolic networks for human liver and Homo sapiens genome • Functional organization of liver revealed by topological modules in liver-specific metabolic network • Enzyme abundance in topological modules of liver-specific metabolic network • Comparison of metabolic network of human liver with that of the Homo sapiens genome Function of liver • producing substances that break down fats • converting glucose to glycogen • producing urea • making certain amino acids • filtering harmful substances from the blood • storing vitamins and minerals • maintaining a proper level of glucose in the blood Network representation of Metabolism: Substrate graph HLPP: The Human Liver Proteome Project The first initiative on human tissues/organs launched by the Human Proteome Organization (HUPO) Data used in this study • Data from CNHLPP Liver metabolic network 6788 distinct proteins (IPI codes) and protein quantitation data , confidence level 95% 6220 distinct genes 1421 genes encode 721 distinct enzymes • BiGG database Human metabolic network 3311 reactions 1555 enzyme-catalyzed reactions 1756 auto-catalytic reactions Reconstruction of liver metabolic network CNHLPP data BiGG 380 enzymes, 1047 enzyme-catalyzed reactions original core reaction set : 1047 liver enzyme-catalyzed reactions initial candidate reaction set: all of the auto-catalytic reactions 2. Extract all metabolites appearing in core reaction set to get core metabolite set. 3. Scan the list of candidate reactions for core metabolites. If all substrates for one reaction can be found in core metabolite set, add this reaction into core reaction set and remove it from the candidate set. 4. If step 3 cannot add any more reactions into core reaction set, stop; else, go to step 2. 1. Added: 427 auto-catalytic reactions Basic graph metrics of metabolic networks Network for human liver Metabolic network Network for H.sapiens genome Nodes 1093 1473 Arcs 2209 3361 Density 0.0019 0.0016 Degree distribution P(k)~k-2.73 P(k)~k-2.67 Average path length 8.5 9.8 Diameter 28 49 Nodes 1026 1407 Arcs 2159 3314 GSC 424 (41.3%) 987 (70.2%) S 262 (25.5%) 117 (8.32%) P 187 (18.2%) 272 (19.3%) IS 153 (14.9%) 31 (2.2%) Biggest cluster Bowtie of biggest cluster Comparison of the liver metabolic network with its random counterparts Liver network Fraction of Diam nodes in eter the biggest cluster Nodes Arcs Densit y Average path length 1093 2209 0.0019 8.5 28 0.9387 100 type I random sub-networks (same arcs as the liver network) Mean 1317 2209 0.0013 10.1 29.7 0.8575 Zscore -21.83 - 29.03 -3.27 -0.47 5.24 100 type II random sub-networks (same nodes as the liver network) Mean 1093 1870 0.0016 9.6 30.2 0.8309 Zscore - 5.25 5.25 -1.25 -0.39 5.09 100 type III random sub-networks (same enzymecatalyzed reactions as the liver network) Mean 1240 2287 0.0015 8.95 26.3 0.8806 Zscore -23.2 -6.17 27.2 -1.8 0.67 5.04 Functional organization of liver revealed by topological modules in liver metabolic network Core-periphery organization Main derivative metabolism functions of the topological modules for human liver-specific metabolic network Module Main Function Main Function category Glycan biosynthesis and metabolism Metabolism of cofactors and vitamins Xenobiotics biodegradation 5 Biosynthesis of chondroitin / heparan sulfate and keratan sulfate 13 Biosynthesis of N-glycan 12 Degradation of heparan sulfate and N-glycan 14 Degradation of chondroitin sulfate 16 Degradation of keratan sulfate 1 Metabolism of folate and vitamin B6 3 R group synthesis 6 Heme biosynthesis 9 Heme degradation and vitamin A metabolism 11 Tetrahydrobiopterin; Vitamin B12 Metabolism 2 ROS detoxification 5 CYP metabolism Enzyme abundance in topological modules of liver-specific metabolic network P (Q >2.35)=10%; P (Q <0.5)=70%. Comparison of metabolic network of human liver with that of the Homo sapiens genome P-value k 1 p 1 i 0 K N K k 1 i n i f (i ) 1 N i 0 n 10 of the 16 modules ( Module 1,2,3,4,6,8,9,11,12,13) : P-value < 0.05 Quantitative difference between categories: overlap score Prototypical overlap score XY (x, y) X (x) Y (y) x X y Y Normalized overlap score XY XY max( XX , YY ) X ,Y : two categorizations X(x) ,Y(y) : the fraction of metabolites in category x X, y Y, respectively XY(x,y): the joint frequency of x and y, i.e. the fraction of vertices that are categorized both as x X and y Y. Quantative difference between a feature of the real metabolic network and its randomized counterparts: Z- score Z f fr f r f : the metric of the feature in the real network fr : the mean of the corresponding metric in the randomized ensemble f r : the standard deviation of the corresponding metric in the randomized ensemble v = 0.72; Z =68.5 Acknowledgement Shanghai Center for Bioinformation and Technology: Lin Tao, Duanfeng Zhang, Kailin Tang ,Ruixin Zhu , Hong Yu ,Yixue Li, Zhiwei Cao Beijing Proteome Research Center: Chao Geng, Ying Jiang,Fuchu He Second Military Medical University: Weidong Zhang Petter Holme Eytan Ruppin Livnat Jerby Ori Folger National Natural Science Foundation of China (10971227, 30900832) Ministry of Science and Technology China(2006AA02312, 2009zx10004601, 2010CB833601 Shanghai Municipal Education Commission (2000236018). Thanks!