- ChemAxon
advertisement

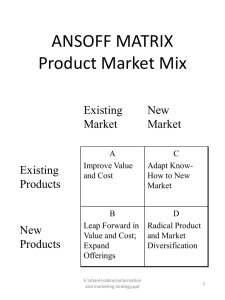
Scientific & technical Presentation Pipeline Pilot Integration Szilárd Dóránt September 2014, Version 14.9.1 The Component Collection: Quick facts • Provides access to ChemAxon tools from Pipeline Pilot • Developed and directly supported by ChemAxon • The component collection itself is free of charge • But still needs the corresponding ChemAxon licenses for the tools being used • Compatibility: • • Each version is compatible only with exact same JChem version since 14.7.7.0 Pipeline Pilot 8.5 or newer required Available functionality (1/3) • Standardizer: structure canonicalization • Chemical Terms expressions for filtering and calculations (including logP, logD, pKa, HBD, HBA, Isoelectric point, PSA and more) • Reactor : smart virtual reaction processing • Maximum Common Substructure (MCS) based clustering • Structural Search and Formula Search filters • Structure Checker Available functionality (2/3) • Name to Structure; Structure to Name; Document to Structure conversion • JChem Base chemical database: insertion, search and retrieval of structures; create and drop structure tables • JChem for Excel export • Marvin applets: structure visualization and editing • Major microspecies (major protonation form) • Microspecies distribution • Burden eigenvalue descriptor (BCUT) Available functionality (3/3) • MolConverter: conversion of the wide range of structure formats supported by ChemAxon • Markush (generic structure) enumeration • Tautomerization: tautomer generation (all, dominant, major, canonical, generic) • Conformer generation • Image generation • RECAP fragmentation Calculator Easy access for the most important calculations More on Calculator plugins Chemical Terms Calculator Maximum freedom trough Chemical Terms Expressions for the expert user • Use arbitrary Chemical Terms expressions • Results stored to arbitrary properties • A wide range of ChemAxon functionality can be accessed as Chemical Terms functions More on Chemical Terms Chemical Terms Filter Filtering with powerful Chemical Terms expressions More on Chemical Terms Standardizer Flexible transformation / canonicalization engine Easy to use, but expert configurations are also accessible: • Simple actions (checkboxes) • Configuration string (simple or XML) • Configuration file More on Standardizer Structure Checker Automated checking and fixing of structures • Pipeline Pilot molecule or structure source input • File or simple action string configuration • Fix or check-only modes • OCR error structures can be ignored • Detected issues, applied fixes and remaining issues are listed in the output More on Structure Checker JChem for Excel Writer Exports live structures to Excel • Pipeline Pilot molecule or structure source input • File output • Export format is Excel 2007 (.xlsx) • Data fields (data record properties) are also exported • Overwrite / append option • Various formatting options More on JChem for Excel Reactor Virtual reaction processing • Supports smart reaction rules to produce synthetically feasible products • Sequential or combinatorial mode • Product or reaction output • Select products to include in output • Use tagger components to distinguish inputs of multi-reactant reactions • Synthesis code generation • Output reaction mapping • Advanced options: – – Unambiguous only Ignore rules: • • • Reactivity and Exclude Selectivity Tolerance More on Reactor Combinatorial Reactor Example Naming components Structure to name and name to structure conversion Example “roundtrip” protocol: More on name recognition Document to Structure Structure extraction from documents • Recognizes • IUPAC and other systematic names • Common names • SMILES, InChi, CAS numbers etc. • OLE objects (“live” structures) • Supports PDF, TXT, Microsoft Office documents, HTML, XML files and URLs • Support for 3 optical structure recognition tools: CLiDE, OSRA, Imago • Correction of some OCR errors • Start page, end page, OSR filtering options • Output: molecule, name, uncorrected name, page number, position, type, OSR confidence More on name recognition Structure Search filter • Substructure, Superstructure, Duplicate, Full Fragment search • Extensive set of search options • Hit highlighting • Support for searching Markush structures JChem Query Guide Formula Search filter • Input types: • Molecule • Formula string • Molecule source • Search types • Exact formula • Exact subformula • Subformula • Support for • Ranges • Multicomponent formula search • Isotopes More on sophisticated chemical formula search Clustering with LibMCS Maximum Common Substructure (MCS) based clustering Options: • Size of smallest common substructure to consider • Three levels of heuristics: – Exact (no heuristics) – Fast – Very Fast • Bond type, atom type, charge, radicals, isotopes can optionally be ignored • Disallow “breaking” rings (default) More on LibMCS Markush Enumeration Enumeration of generic structures • File input • Enumeration type: – Sequential – Random • Number of enumerated structures can be limited (per input structure) • Valence filter • Scaffold alignment • Markush code generation. The scaffold ID can be: – fetched from data field – generated (prefix + number) More on Markush Enumeration Tautomerization Component for tautomer generation • Calculation modes: – All tautomers – Canonical tautomer – Generic tautomer – Major tautomer – Dominant tautomer distribution • Options: – Protect aromaticity, charge, double bond stereo, tetrahedral stereo – Exclude antiaromatic compounds – Single fragment mode – Consider pH at specific value More on Tautomerization Conformer generation Component for 3D conformer generation • Calculation modes: – Multiple conformers – Lowes energy conformer • Options: – Maximum number of conformers – Diversity limit – Optimization limit, hyperfine option – Time limit – Generate with explicit H atoms – Energy unit kcal/mol or KJ/mol, into arbitrary property More on conformer generation MolConverter “Swiss army knife” for molecular format conversion • Input and output can either be – File – Property – Pipeline Pilot Molecule • Specified input format or autodetection • Various output formats or custom format string • Option to halt or continue on error, error messages put into property • 2D cleaning (coordinate generation) only when needed (default). Unconditional 2D or 3D cleaning or no cleaning can also be selected More on supported file formats Fragmenter RECAP based fragmentation • Molecule fragmentation based on predefined cleavage rules • Support for marking attachment points - As any (*) atoms - As Al and Ar atoms for aliphatic and aromatic distinction • Cut data can be added as atom labels • Detailed cleavage data is stored in properties More on Fragmenter Image Generation High-quality ChemAxon-rendered images • Image formats: PNG, BMP, JPEG • Input can be either - Pipeline Pilot Molecule - Structure source (e.g. MRV string) • Numerous rendering options, for example: - Image size, background, transparency - Scaling, max scale, atom label size - Various aromatization, dearomatization modes - R/S label, E/Z label, Absolute label options - Mark valence errors - Implicit H display, add/remove explicit H - etc … HTML Molecular Spreadsheet Scalable molecule and data display • Adds ChemAxon display capabilities to the familiar “HTML Molecular Table Viewer” Pipeline Pilot component • Supports ChemAxon hit coloring, advanced Markush features • Larger image pop-up • Applet pop-up • Wide array of display options More on MarvinView HTML Molecular Spreadsheet More on MarvinView Database Connection • Provides a convenient way to define a JDBC connection parameter set within a protocol • Other JChem Base components refer to this parameter set by a symbolic name (e.g. “myConnection”) • Multiple instances may be used in a protocol if needed • Each component creates its own JDBC connection to the database according to these parameters JChem Base table creation Creates a JChem Base table • Different table types supported • Non-default fingerprint parameters can be specified • Absolute Stereo Flag option • Duplicate filtering option • Tautomer duplicate filtering • Custom Standardizer configuration can be specified • Extra column definitions can be added as SQL suffix More on JChem Base JChem Base Insert Inserts structures into a JChem Base table • Duplicate filtering uses Pass and Fail ports if set • Returns cd_id (primary key) values • Two input modes: – read structure source from a specified property – if property not specified uses Pipeline Pilot input molecule • Insert into additional data fields • Option to continue on error, error message stored in specified property More on JChem Base JChem Database Search (1/2) Search in a JChem Base table • An extensive number of search options supported JChem Query Guide JChem Database Search (2/2) Highlighted component features: • Modes of operation: - Hit return mode - Flow-trough (“Query filtering”) mode • Various output options for DB hits: - cd_id value (primary key) - Pipeline Pilot molecule - Generated MRV source or original source from DB • Hit coloring supported Hit alignment - Rotate - Partial clean • Markush hit reduction supported (with MRV output) • Option for fetching data fields from JChem Base structure table Delete from JChem Base table Deletes rows from a JChem Base table • Delete by input list of cd_id (primary key) values, for example results of a search operation • Delete by SQL WHERE clause, e.g. “WHERE cd_id IN (23, 247, 786)” • Delete all rows by empty WHERE clause More on JChem Base JChem Base demo protocol Resources • Download: – http://www.chemaxon.com/download/pipeline-pilotcomponents • Technical support forum: – http://www.chemaxon.com/forum/forum88.html • E-mail: – pp-support@chemaxon.com • More resources: – http://www.chemaxon.com/forum/ftopic4604.html Visit other technical presentations MarvinSketch/View http://www.chemaxon.com/MarvinSketch_View.ppt MarvinSpace http://www.chemaxon.com/MarvinSpace.ppt Calculator Plugins http://www.chemaxon.com/Calculator_Plugins.ppt JChem Base http://www.chemaxon.com/JChem_Base.ppt JChem Cartridge http://www.chemaxon.com/JChem_Cartridge.ppt Standardizer http://www.chemaxon.com/Standardizer.ppt Screen http://www.chemaxon.com/Screen.ppt JKlustor http://www.chemaxon.com/JKlustor.ppt Fragmenter http://www.chemaxon.com/Fragmenter.ppt Reactor http://www.chemaxon.com/Reactor.ppt