Data_Analsis
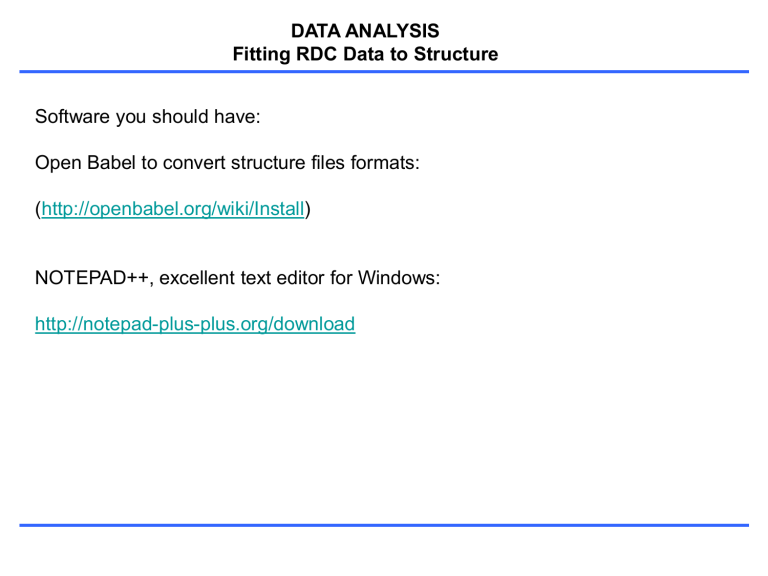
DATA ANALYSIS
Fitting RDC Data to Structure
Software you should have:
Open Babel to convert structure files formats:
( http://openbabel.org/wiki/Install )
NOTEPAD++, excellent text editor for Windows: http://notepad-plus-plus.org/download
DATA ANALYSIS
Fitting RDC Data to Structure
Software for performing SVD fitting of structure to RDCs data:
MSpin by Armando Navarro-Vazquez, Commercialized by Mestrelab Research,
Santiago de Compostela, SPAIN.
http://mestrelab.com/software/mspin/
PALES by Markus Zweckstetter, Max Plank Institute for Biophysical Chemistry,
Göttingen, GERMANY.
http://www.mpibpc.mpg.de/groups/zweckstetter/_links/software_pales.htm
DATA ANALYSIS
Fitting RDC Data to Structure
In order to fit the data to a set of judicious structures (all the configurational space of your molecule) you need to know more than five independent (non-parallel internuclear vector) RDCs.
At least three of them have to be out of the plane.
You need a minimun of five RDCs to calculate the alignment tensor ( A ), but need more than five to perform the fitting using Singular Value Decomposition analysis either with the programs MSpin or PALES.
Each fitting will give you a quality factor Q (Cornilescu Q factor J. Am. Chem.
Soc. 1998, 120, 6836-6837). The lower the Q factor the better the fitting.
Fitting structures in MSpin is very straightforward. You can use the tutorial from
Mestre website. MSpin reads any type of PDB files and also the XYZ format.
On the other side, PALES follows a unique PDB file format.
PALES file format
The structure file:
For Small Molecules, PALES reads a structure file that is an adaptation of the original
PDB file format used for proteins. Instead of having N number of residues
(e.g. aminoacids), the file represents a single residue (N=1).
PRACTICAL EXAMPLES
LUDARTIN
14
O
2
3
4
15
1
5
H
O
6
10
9
7
8
11
13
O
1
12
O
H
O
2
O
- gastric cytoprotective effect
- inhibits the aromatase enzyme
- first isolated in 1972 from Artemisia carruthii by Geissman and Griffin as a mixture with it 11,13-dihydroderivative
- the stereochemistry displayed in 1 based on the chemical shift and coupling constants of H-6 and on the chemical shift of H-15
Giordano, O. S.; Guerreiro, E.; Pestchanker, M. J.; Guzman, J.; Pastor, D.; Guardia, T. J. Nat. Prod. 1990, 53, 803-9.
Blanco, J. G.; Gil, R. R.; Alvarez, C. I.; Patrito, L. C.; Genti-Raimondi, S.; Flury, A. FEBS Lett. 1997, 409, 396-400.
- Geissman, T. A.; Griffin, T. S. Phytochemistry 1972, 11, 833-5.
Determination of the Stereochemistry of Ludartin ( 1 ) Using Chemical Transformations
14
O
2
3
4
15
1
5
H
O
6
10
9
7
8
11
13
O
1
12
H
O
2
O
O
H
O
3
O
+
O
H
O
4
O
Sosa, V. E.; Oberti, J. C.; Gil, R. R.; Ruveda, E. A.; Goedken, V. L.; Gutierrez, A. B.; Herz,
W. Phytochemistry 1989 , 28 , 1925-9.
H-3
H-3 and CH
3
-15
27 o above and below the plane of the five-membered ring
CH
3
-15
RMS fit and overlay of the 3D structures of ludartin ( 1 ) and 3,4-b-epoxyludartin ( 2 ) using only the heavy atoms belonging to the 5-member and the 7-member rings, and the lactone ring.
RMS error: 0.039 Å
PALES Presentation
This is the methylacetamide PDB file created by HyperChem
PALES can not read this file. It does not recognize HETATM
HETATM 1 C 1 -1.413 -1.626 0.000
HETATM 2 C 2 -1.342 -0.119 0.000
HETATM 3 N 3 -2.536 0.577 -0.000
HETATM 4 C 4 -2.573 2.004 0.000
HETATM 5 O 5 -0.252 0.487 -0.000
HETATM 6 H 6 -0.373 -2.033 -0.000
HETATM 7 H 7 -1.947 -1.993 0.910
HETATM 8 H 8 -1.947 -1.993 -0.910
HETATM 9 H 9 -3.390 0.079 0.000
HETATM 10 H 10 -3.637 2.354 -0.000
HETATM 11 H 11 -2.048 2.408 0.908
HETATM 12 H 12 -2.048 2.408 -0.908
CONECT 1 2 6 7 8
CONECT 2 1 3 5
CONECT 3 2 4 9
CONECT 4 3 10 11 12
CONECT 5 2
CONECT 6 1
CONECT 7 1
CONECT 8 1
CONECT 9 3
CONECT 10 4
CONECT 11 4
CONECT 12 4
END
Save the structure generate by HyperChem in hin format
Convert the hin format to a PDB file using Open Babel
PDB file created by Open Babel from hin file
COMPND C:\Users\rgil\SMASH 2010 RDCs Workshop\Methyl Acetamide\methylacetamide.hin
AUTHOR GENERATED BY OPEN BABEL 2.2.3
HETATM 1 C LIG 1 -1.413 -1.626 0.000 1.00 0.00 C
HETATM 2 C LIG 1 -1.342 -0.119 0.000 1.00 0.00 C
HETATM 3 N LIG 1 -2.536 0.577 -0.000 1.00 0.00 N
HETATM 4 C LIG 1 -2.573 2.004 0.000 1.00 0.00 C
HETATM 5 O LIG 1 -0.252 0.487 -0.000 1.00 0.00 O
HETATM 6 H LIG 1 -0.373 -2.033 -0.000 1.00 0.00 H
HETATM 7 H LIG 1 -1.947 -1.993 0.910 1.00 0.00 H
HETATM 8 H LIG 1 -1.947 -1.993 -0.910 1.00 0.00 H
HETATM 9 H LIG 1 -3.390 0.079 0.000 1.00 0.00 H
HETATM 10 H LIG 1 -3.637 2.354 -0.000 1.00 0.00 H
HETATM 11 H LIG 1 -2.048 2.408 0.908 1.00 0.00 H
HETATM 12 H LIG 1 -2.048 2.408 -0.908 1.00 0.00 H
CONECT 1 2 6 7 8
CONECT 2 1 3 5 5
CONECT 3 2 4 9
CONECT 5 2 2 boxes is not used by PALES.
ERASE IT
CONECT 8 1
CONECT 9 3
CONECT 10 4
CONECT 11 4
CONECT 12 4
MASTER 0 0 0 0 0 0 0 0 12 0 12 0
END
Edit the file with a good text editor accordingly. I recommend NOTEPAD++
HETATM 1 C LIG 1 -1.413 -1.626 0.000 1.00 0.00
HETATM 2 C LIG 1 -1.342 -0.119 0.000 1.00 0.00
HETATM 3 N LIG 1 -2.536 0.577 -0.000 1.00 0.00
HETATM 4 C LIG 1 -2.573 2.004 0.000 1.00 0.00
HETATM 5 O LIG 1 -0.252 0.487 -0.000 1.00 0.00
HETATM 6 H LIG 1 -0.373 -2.033 -0.000 1.00 0.00
HETATM 7 H LIG 1 -1.947 -1.993 0.910 1.00 0.00
HETATM 8 H LIG 1 -1.947 -1.993 -0.910 1.00 0.00
HETATM 9 H LIG 1 -3.390 0.079 0.000 1.00 0.00
HETATM 10 H LIG 1 -3.637 2.354 -0.000 1.00 0.00
HETATM 11 H LIG 1 -2.048 2.408 0.908 1.00 0.00
HETATM 12 H LIG 1 -2.048 2.408 -0.908 1.00 0.00
END
PALES is written in C and generally it is very forgiving in terms of format, i.e. it does not care whether you use spaces, tabs, ...
However, editors from windows such as NOTEPAD or WORDPAD may intruduce characters that can make the file unreadeable by PALES. This not always the case but it may happen.
The only requirement is that the naming convention in the PDB file and the RDC table are identical. In addition, PALES only takes into account lines in the PDB file starting with "ATOM". For the MAC and LINUX versions of the GUI will also include an automatic reformatting step for the PDB file. Then at least in the GUI these problems should not show up.
Edit the file with a good text editor accordingly
HETATM 1 C LIG 1 -1.413 -1.626 0.000 1.00 0.00
HETATM 2 C LIG 1 -1.342 -0.119 0.000 1.00 0.00
HETATM 3 N LIG 1 -2.536 0.577 -0.000 1.00 0.00
HETATM 4 C LIG 1 -2.573 2.004 0.000 1.00 0.00
HETATM 5 O LIG 1 -0.252 0.487 -0.000 1.00 0.00
HETATM 6 H LIG 1 -0.373 -2.033 -0.000 1.00 0.00
HETATM 7 H LIG 1 -1.947 -1.993 0.910 1.00 0.00
HETATM 8 H LIG 1 -1.947 -1.993 -0.910 1.00 0.00
HETATM 9 H LIG 1 -3.390 0.079 0.000 1.00 0.00
HETATM 10 H LIG 1 -3.637 2.354 -0.000 1.00 0.00
HETATM 11 H LIG 1 -2.048 2.408 0.908 1.00 0.00
HETATM 12 H LIG 1 -2.048 2.408 -0.908 1.00 0.00
END
Editing:
1) Replace all HETATM with ATOM
2) Insert the word TER before END
3) Add the proper number next to each atom label (C, N, O, H, etc)
4) See edited file in next slide
Edit the file with a good text editor accordingly
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
ATOM
TER
END
1 C 1 LIG 1 -1.413 -1.626 0.000 1.00 0.00
2 C 2 LIG 1 -1.342 -0.119 0.000 1.00 0.00
3 N 3 LIG 1 -2.536 0.577 -0.000 1.00 0.00
4 C 4 LIG 1 -2.573 2.004 0.000 1.00 0.00
5 O 5 LIG 1 -0.252 0.487 -0.000 1.00 0.00
6 H 6 LIG 1 -0.373 -2.033 -0.000 1.00 0.00
7 H 7 LIG 1 -1.947 -1.993 0.910 1.00 0.00
8 H 8 LIG 1 -1.947 -1.993 -0.910 1.00 0.00
9 H 9 LIG 1 -3.390 0.079 0.000 1.00 0.00
10 H 10 LIG 1 -3.637 2.354 -0.000 1.00 0.00
11 H 11 LIG 1 -2.048 2.408 0.908 1.00 0.00
12 H 12 LIG 1 -2.048 2.408 -0.908 1.00 0.00
ATOMNAME RESNAME RESID
NOTE: The changes are printed in red
Codes uses by the
PALES RDC input file
See next slide
ATOMNAME is the atom number in the structure file
RESNAME is a fake name to simulate a residue (e.g. Lys, Ala, etc, in a peptide),
LIG in this case, but you can use any name as long as you use de same name in
RDCs input table.
RESID is the number of the residue in peptide sequence, for a small molecule is just 1.
The RDCs table file in PALES
VARS RESID_I RESNAME_I ATOMNAME_I RESID_J RESNAME_J ATOMNAME_J D DD W
FORMAT %5d %6s %6s %5d %6s %6s %9.3f %9.3f %.2f
1 LIG C1 1 LIG H6 -6.800 1.000 1.00
1 LIG C1 1 LIG H5 0.800 1.000 1.00
1 LIG C1 1 LIG H8 -25.230 1.000 1.00
1 LIG N3 1 LIG H9 -15.6 1.000 1.00
1 LIG C4 1 LIG H8 -75.5 1.000 1.00
1 LIG C4 1 LIG H10 7.510 1.000 1.00
1 LIG C4 1 LIG H11 -4.530 1.000 1.00
1 LIG C4 1 LIG H12 123.0 1.000 1.00
RDCs (Hz)
Experimental
Error (Hz)
Fitting data to structure using command line PALES in Windows:
To perform an SVD fitting in PALES you have to execute the following command: pales –bestFit –pdb –name.pdb –inD rdc_file.tab –outD output.SVD.file
name.pdb is the name of the PDB file rdc_file.tab is the name of the RDC data file
The output is very well explained in the PALES documentation or in the Nature
Protocols paper (See below). Right now we are only interested on the Q factor.
The lower the Q factor the better the SVD fitting.
See also:
Nature Protocols 2008 3(4) 679-690
PERSPECTIVES
Future Directions on RDCs in Small Molecules
-Alignment Media: more variety (PEO), more deuteration, chiral gels for all solvents, …
-Low Temperature Alignment Media to resolve conformational average.
Commercialization: gels, stretching apparatus, software, etc…
-Adapted Measurement Techniques: in conjunction with scaling of RDCs novel pulse sequences
-Software: Individual error treatment, incorporation of RCSA, automation of structure generation, ab initio and
MD methods for treating flexible molecules, prediction of alignment (absolute configuration)
-Technical Improvements: Gradient shimming
Revisit Underutilized Experiments
Selective 1D NOESY
NOE buildup curves of H10 while selectively exciting H4 axial of cortisol (1) in DMSO-d6: (a) the
‘‘raw’’ NOE buildup curve; (b) the
NOE buildup curve obtained with
PANIC (peak amplitude normalization for improved crossrelaxation)
Series of 1D NOESY Spectra of Ludartin
H-3 is selectively excited
H-2a H-2b
H-15
600 ms
100 ms
Steps of 100 ms
14
NOE 1/r 6
O
2
3
4
15
1
5
H
O
6
10
9
7
8
11
13
12
O
3.9
3.8
3.7
3.6
3.5
3.4
3.3
3.2
3.1
3.0
2.9
2.8
2.7
2.6
2.5
1
2.4
2.3
2.2
O
2.1
2.0
1.9
1.8
1.7
1.6
1.5
1.4
1.3
1.2
1.1
ppm
H
O
2
O