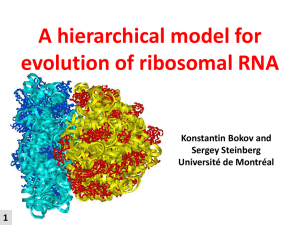
Annotated tertiary interactions in RNA structures reveal new
advertisement

Towards RNA structure prediction:
3D motif prediction and
knowledge-based potential
functions
Christian Laing
Tamar Schlick’s lab
Courant Institute of Mathematical Sciences
Department of Chemistry
New York University
RNA folding is hierarchical
Sequence
Secondary Structure
Annotated diagram
3D Structure
5‘gGACUCG
GGGUGCCC
UUCUGCGU
GAAGGCUG
AGAAAUACC
CGUAUCAC
CUGAUCUG
GAUAAUGC
CAGCGUAG
GGAAGUUc3
'
TPP riboswitch (PDB: 2GDI)
• Tertiary motifs serve as modular building blocks in the
RNA architecture.
• To understand the role of RNA tertiary motifs in RNA
folding will help to understand RNA 3D prediction.
Annotating 3D RNA
• Selected seven key RNA tertiary motifs:
coaxial helix, A-minor motif, ribose zipper,
tetraloop-tetraloop receptor, pseudoknot,
kissing hairpin, and tRNA D-loop:T-loop.
• Searched RNA tertiary motifs via different
computer programs
• Annotate tertiary interaction motifs.
• Perform analysis over the diagrams
produced.
Examples
RNA junctions have a high probability (84%) to
contain at least one coaxial helix.
Distribution of tertiary motifs
tRNA D-loop;T-loop
7 (1%)
Loop-loop receptor
16 (3%)
Kissing hairpin 6
(1%)
Coaxial helix 182
(30%)
Pseudoknot 40 (7%)
Ribose zipper 121
(20%)
A-minor motif 229
(38%)
• For 54 high-resolution RNA structures, 615 RNA tertiary
interactions were found.
• Ribose zippers, coaxial helices and A-minor interactions
are highly abundant (88%).
Correlated motifs
• Several A-minor motifs
(64%) are involved with
coaxial helices.
• Coaxial helices (70%)
interact with A-minor.
• Most ribose zippers (70%)
contain an A-minor.
• Every loop-loop receptor
contains a ribose zipper,
which in turn contains one
or more A-minor motifs 87%
of the time.
Group I intron (PDB: 1HR2)
RNA V.5 1119 (2001)
Can we predict coaxial stacking?
HCV virus fragment (PDB: 1KH6)
NAT.STRUCT.BIOL. V. 9 370 2002
Topology of 4-way junctions in folded RNAs
Family H
Family K
Analysis on 24 junctions:
GUIDELINES
• A coaxial staking Hi-Hi+1 is
enhanced when Ji,i+1 is small.
• There is a strong preference
for a H1-H4 stacking.
• Coaxial helices in family H
have similar lengths.
• Pseudoknots usually stack
their helices.
Family X
Family t
Family
<J12>
<J23>
<J34>
<J41>
Coaxial H.
H: 10
H: 1
2.0
0
0.5
0
2.4
0
0.0
0
H1-H4; H2-H4
H1-H2; H3-H4
K: 3
K: 4
K: 1
5.0
4.3
2
2.3
3.8
0
4.3
1.8
1
0.3
1.3
0
H1-H4
H3-H4
H2-H3
X: 5
4.4
2.2
5.6
3.8
t: 1
6
4
5
1
H2-H4
A-minor involved in long-range interactions
Structural context of the inserted A in A-minor
Structural context of the Watson-Crick pair in Aminor
40
35
50
25
Percentage
Percentage
30
20
15
10
5
40
30
20
10
0
0
Helix (WC)
Internal
Terminal
Structural context
Junction
Other SS
1
2
3
Helical context
4
5
Statistical Potentials for A-minor prediction
• The adenosine (A) can be located in four
single stranded regions: internal (I) and
terminal (T) loops, junctions (J), and other (O).
• The helix receptor (R) can be located in five
positions relative to the end site of a helix.
• The potential function for an A-minor ( Ai , Rj ) is defined by:
N Obs ( Ai , R j )
P( Ai , R j )
,
N Exp ( Ai , R j )
Ai {AI , AT , AJ , AO },
Rj {R1, R2 , R3 , R4 , R5}
• NObs ( Ai , R j ) is the observed number of interacting pairs ( Ai , Rj ).
• NExp ( Ai , R j ) is the expected number of interacting pairs ( Ai , R j ).
• The statistical free energy for each A-minor is:
G( Ai , Rj ) kBT ln(P( Ai , R j ))
kB Boltzmann const., T Temperature
Improving coarse-grained model
Rosetta:
- 1-bead model
(only the base).
- Neglect sugar and
phosphate.
- Only one RNA (23S
rRNA) was considered.
PNAS V. 104 No. 37 14664-9 (2007)
Possible improvements:
- 3-bead model to consider
base, sugar, and
phosphate.
- Consider our 54-nonredundant RNA dataset.
PNAS V. 102 No. 19 6789-94 (2005)
From 2D to 3D: RNA assemble
Assume we know the RNA secondary structure (and
possible some constrains), how can we use this knowledge
into our Mesoscopic model?
• Stems can be described by type-A helices.
• Programs such as Assemble (Westhof) and RNA2D3D
(Shapiro) can be used but require manual intervention.
Suggested future directions
• Design statistical potentials for coaxial stacking.
• The combination of A-minor/coaxial helix
prediction may lead to stronger arguments.
• Design statistical potentials to predict non
canonical basepairs (Leontis/Westhof), and
explore the possibility to use them with dynamic
programming.
• Test the statistical potentials (threading, decoys).
• There are 272 non-redundant 4-way junctions in
the RNA junction database that can be analyzed
topologically and geometrically.
Acknowledgements
Yurong Xin and Tamar Schlick
Many thanks to:
Hin Hark Gan
Sean McGuffee
Shereef Elmetwaly
Human Frontier
Science Program
NSF/NIGMS initiative
in Mathematical
Biology (DMS-0201160)
Topology of 3-way junctions in folded RNAs
(Lescoute & Westhof RNA 2006 12: 83-93)
Analysis over 33 junctions.
Back up slide
Back up slide
Back up slide