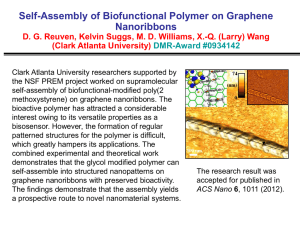
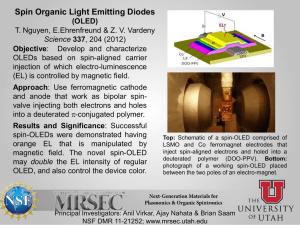
Translocation
advertisement

Anomalous Dynamics of Translocation Mehran Kardar MIT COLLABORATORS Yacov Kantor, Tel Aviv Jeffrey Chuang, UCSF Supported by OUTLINE Polymer dynamics – biological examples and technological applications Translocation as an “escape” problem Anomalous dynamics of free translocating polymers Translocation under influence of force Conclusions Dynamics of polymers in confined geometries Accumulation of exogenous DNA in host cell nucleus: • viral infection • gene therapy • direct DNA vaccinations Motion of DNA through a pore can be used to read-off the sequence Motion of polymer in random environments DNA gel electrophoresis or reptation A reconstituted nucleus being dragged after a 3-µm-diameter bead, linked by a molecule of DNA. The time interval between measurements in the first and second images is 532 sec, between the second and third, 302 sec. Note the shortening of the maximum distance between bead and nucleus. Salman, H. et al. (2001) Proc. Natl. Acad. Sci. USA 98, 7247-7252 What is a pore in a membrane? Alpha-hemolysin secreted by the human pathogen Staphylococcus aureus is a 33.2kD protein (monomer); It forms 232.4kD heptameric pore Song, Hobaugh, Shustak, Cheley, Bayley, Gouaux Science 274, 1859 (1996) Measuring translocation of a polymer Single-stranded DNA molecules (negatively charged) are electrically driven through a pore Meller, Nivon, Branton PRL 86, 3435 (2001) Measuring translocation of a polymer (cont’d) Voltage driven translocation Method of measuring translocation times “in the absence of driving force” Bates, Burns, Meller Biophys.J., 84,2366 (2003) Translocation through a solid membrane Can we use translocation to read-off a DNA sequence? (“B”-real trace; “C”- “cartoon”) Computer simulations of complicated problems Trapped polymer chain in porous media Baumgartner, Muthukumar, JCP 87, 3082 (1987) Polymer escape from a spherical cavity N=60, t=50,350,450,1000,4850,25000 Muthukumar, PRL 86, 3188 (2001) “Translocation” – the simplest problem Find mean translocation time & its distribution as a function of N, forces, properties of the pore Entropy of “translocating” polymer N-s s Reviews: Eisenriegler, Kremer, Binder JCP 77, 6296 (1982); De Bell, Lookman RPM 65, 87 (1993) Diffusion over a barrier – Kramers’ problem p V k BT s m in H.A. Kramers, Physica 7, 284 (1940) s max s Is there a “well” in the entropic problem? Free energy for N=1000 as a function of translocation coordinate s Chuang, Kantor, Kardar, PRE 65, 011892 (2001) Sung, Park, PRL 77, 783 (1996); Muthukumar, JCP 111, 10371 (1999) Is there a “well” in the entropic problem? (contn’d) Distribution of escape times with (dashed) and without (solid) barrier Chuang, Kantor, Kardar, PRE 65, 011892 (2001) Smoluchowski equation vs. simulation the case of 3D phantom chain Distribution of translocation coordinate n for 3 different times (N=100); continuous lines represent fitted solutions of Smoluchowski equation (D=0.011) S.-S. Chern, A.E. Cardenas, R.D. Coalson JCP 115, 7772 (2001) Translocation vs. free diffusion Translocation is faster than free diffusion!??? Monte Carlo model min=2, max=101/2 Carmesin, Kremer Macromol.21, 2819 (1988) 1D phantom polymer model max=2, “w=1” “w=3” Translocation time of 1D phantom polymer Distribution of translocation times of 1D phantom polymers (normalized to mean) averaged over 10,000 cases (N=32,64,128) vs. solution of FP equation Translocation time of 1D phantom polymers averaged over 10,000 cases Chuang, Kantor, Kardar, PRE 65, 011892 (2001) Scaled translocation times of 1D phantom polymers as a function pf pore width w averaged ofver10,000 cases (N=3,4,6,8,128,181) Translocation time of 2D polymer Ratio between translocation times of 2D phantom and selfavoiding polymers with and without membrane Translocation time of 2D phantom & self-avoiding polymers (averaged over 10,000 cases) Effective exponents for 2D phantom and selfavoiding polymers with and without mebrane Note: in d=2, 1+2n=2.5 Chuang, Kantor, Kardar, PRE 65, 011892 (2001) Anomalous diffusion of a momomer Kremer, Binder, JCP 81, 6381 (84); Grest, Kremer, PR A33, 3628 (86); Carmesin, Kremer, Macromol. 21, 2819 (88) Anomalous translocation of a polymer Time dependence of fluctuations in translocation coordinate in 2D self-avoiding polymer. The slope approaches 0.80. Y. Kantor and M. Kardar, Phys. Rev. E 69, 021806 (2002) Translocation with a force applied at the end Distribution of translocation times for N=128and values of Fa//kT=0, 0.25 and infinity, for 2D self-avoiding polymer. 250 configurations. Scaled inverse translocation time in 2D self-avoiding polymer as a function of scaled force. Kantor, Kardar (2002) “Infinite” force applied at the end Translocation of 2d self-avoiding polymer under influence of infinite force at t=0, 60,000, 120,000 MC time units Scaled inverse translocation time in 2D self-avoiding polymer as a function of N under influence of an infinite force. Slope of the line is 1.875. Kantor, Kardar (2002) “Infinite” force applied to phantom polymer “Snapshots” of spatial configuration of translocating 1D phantom polymer (N=128) under influence of infinite force at several stages of the process Translocation time of 1D phantom polymer as a function of N under influence of an infinite force (circles) and motion without membrane (squares). Slopes of the lines converge to 2.00 [Kantor, Kardar (2002)] “Infinite” force applied to free phantom polymer “Snapshots” of spatial configuration of 1D phantom polymer (N=128) moving under influence of infinite force. The position of first monomer was displaced to x=0. Kantor, Kardar (2002) Short time scaling Position of the first monomer of 1D phantom polymer as a function of scaled time in the absence of membrane for N=8,16,32,…,512. Position of the first monomer of 1D phantom polymer as a function of scaled time during the translocation process for N=8,16,32,…,512. Kantor, Kardar (2002) “Infinite” CPD – phantom polymer Translocation time in 2D phantom polymer as a function of N under influence of an infinite chemical potential difference. Slope of the line is 1.45. Kantor, Kardar (2002) Translocation with chemical potential difference Distribution of translocation times for N=64and values of U/kT=0, 0.25, 0.75 and 2, for 2D self-avoiding polymer. 250 configurations. Scaled inverse translocation time in 2D self-avoiding polymer as a function of scaled U . Kantor, Kardar (2002) “Infinite” chemical potential difference Translocation of 2D self-avoiding polymer under influence of infinite chemical potential difference at t= 10,000, 25,000 MC time units. Kantor, Kardar (2002) Translocation time in 2D self-avoiding polymer as a function of N under influence of an infinite chemical potential difference. Slope of the line is 1.53. Conclusions/Perspectives Normal diffusion “explains” only Gaussian polymers and gives “wrong” prefactors Anomalous dynamics provides a consistent picture of translocation There is no detailed theory that will enable calculation of coefficients Crossovers persist even for N~1000. We presented “bounds” for diffusion under influence of large forces. Are they the “real answer”?