mtm4
advertisement
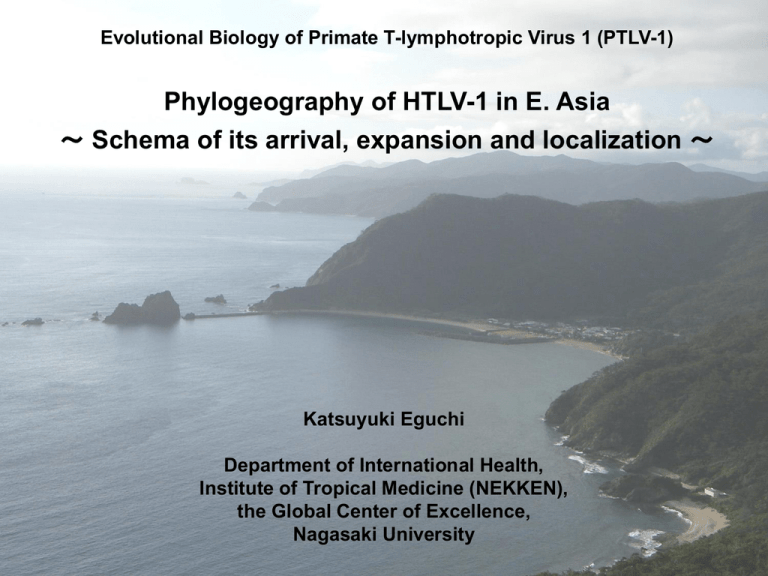
Evolutional Biology of Primate T-lymphotropic Virus 1 (PTLV-1) Phylogeography of HTLV-1 in E. Asia ~ Schema of its arrival, expansion and localization ~ Katsuyuki Eguchi Department of International Health, Institute of Tropical Medicine (NEKKEN), the Global Center of Excellence, Nagasaki University Background HTLV-1 was discovered as an agent of Adult T-Cell Leukemia. Ten to 20 million people worldwide are infected with HTLV-1, although the aggregations of the carriers are seen in Western Africa, Papua New Guinea, part of South America and Japan. Verdonck et.al. 2007 Even within Japan unique distribution of the carriers is found. The southwestern Japan is the area with the highest HTLV-1 prevalence in the world. In early 1980’s more than 30% of the population in Goto Is. (Nagasaki Pref.) was infected. But, today, the dramatic decline of HTLV-1 prevalence is reported in Japan. Tajima: in Hinuma & Sakitani (ed) 2003 HTLV-1 is genetically close to HIV but there are several differences: HTLV-1 was probably derived from simian retrovirus more than 10,000 years ago, while HIV can be traced back as far as 1950’s. HTLV-1 predominantly transmits vertically through breast-feeding from mother to child, while HIV spread horizontally mainly through sexual contacts. Only a few percents of HTLV-1 carriers develop ATL (Adult T-cell Leukemia) more than 50 years after infection, while 95% of HIV carriers without any treatment cause AIDS within 10 years on average after infection. This may indicate that HTLV-1 stand up at a late stage of the co-evolution with human being. Or, HTLV-1 now faces “climax” at the end of the co-evolution, because the prevalence of HTLV-1 has recently declined naturally and dramatically in Japan. Implications to study a long-term dynamics of HTLV-1 Through this study we will have a new insight to understand natural history of human retrovirus infection, and co-evolution of human and human retroviruses and the subsequent impacts to human societies. Research Questions at the beginning When did the HTLV-1 arrive at Japan? Who carried the HTLV-1 into Japan? Prehistoric hypothesis = Introduction in prehistoric age by the earlier settlers. European Trade hypothesis = Introduction by Africans accompanying European traders several centuries ago (from 16th to 17th century). How has the HTLV-1 expanded and localized inside Japan? This kind of study may be referred to as “Phylogeography”: Science to study temporal and spatial distribution of certain organisms and to detect biological and environmental factors driving such dynamics. Approaches Finding the ratio of Transcontinental subgroup (TC) to Japanese subgroup (JPN) in endemic areas within East Asia. HTLV-1 is subdivided into 7 subtypes: Subtype A: cosmopolitan, consisting of five subgroups: A, Transcontinental B, Japanese C, West African D, North American E, Peruvian Subtype B: Central African Subtype C: Melanesian Subtype D: Central African Subtype E: D. R. Congo Subtype F: Gabon Subtype G: Central and West African Transcontinental subgroup (TC) and Japanese subgroup (JPN) of the cosmopolitan subtype are found in Japan and adjacent areas. Reconstructing molecular phylogeny of East Asian isolates of HTLV-1. Study Area We conducted field surveys in the Kakeroma Island of the Amami Islands. Historically, the Amami Islands are a site of migration for Ryukyuans and Wajin and there has been considerable socio-cultural interchange between them. Methods (1) Extracting DNA from 23 carriers residing in Kakeroma island, N. Ryukyus DNA of 197 HTLV-1 carriers provided from the Joint Study on Predisposing Factors of ATL Development (JSPFAD) Hokkaido Iwate Kochi Hirado Kumamoto Okinawa Amplifying env region by PCR Genotyping isolates by RFLP (Hpa I, Hinf I) TC JPN 40 4 30 50 23 50 Methods (2) Extracting DNA from 23 carriers residing in Kakeroma island, and 5 in Ishigaki Amplifying 5’LTR region by PCR Sequencing ca. 650 bp fragment. Reconstructing phylogenetic trees DNA of 197 HTLV-1 carriers provided from the Joint Study on Predisposing Factors of ATL Development (JSPFAD) Results (1): Distribution of JPN and TC in Japan A “centrifugal” pattern may be detected. Results (2): Phylogenetic Analysis This is a phylogenetic tree of HTLV-1 drawn by “Bayesian method”. Frankly speaking, this is a “family tree” of the virus. The isolates highlighted in pink corresponded to JPN, those in blue to TC. This tree clearly shows that TC is an ingroup of JPN Discussions Two hypotheses of the origin of HTLV-1 have been proposed: Prehistoric hypothesis European Trade hypothesis European Trade hypothesis is unlikely because: The two major international seaports at that time are Hirado and Nagasaki where JPN is prevalent, but JPN has so far been known almost exclusively from Japan. TC is known from Ainu people and Ryukyuans. Although TC is also found from Africa, little direct contacts between Africans and Ainu people living in the northern extreme of Japan must have been taken place in 16th and 17th centuries. Our results support Prehistric hypothesis. A possible scenario creating the present distribution of TC and JPN is as follows. Before or in the initial new stone age the earliest settlers (mt haplotypes C1and C3?) came to Japan and spread over the archipelago. They may brought TC to Japan. In initial Jomon period the second weave of immigrants (D2?) arrived at Kyushu through the Korean Peninsular, and then spread northeastward and southwestward. They may brought JPN to Japan. In Yayoi and Kofun period the weaves of immigrants (O2b and O3) arrived at Western Japan, and then their population increased rapidly and resulted in the establishment of their ancient nation(s). They may be non-carriers, and, due to their rapid population growth, endemic areas of HTLV-1 gradually disappear from the central part of Japan. Summary of our findings Our results support the Prehistoric hypothesis. JPN (almost exclusively known from Japan) was not derived from the widespread TC. Actually, TC was recognized as an ingroup of JPN. This may suggest that JPN is not originated from Japan but both JPN and TC were originated in the continental Asia and then introduced into Japan. In order to explain the present distribution of HTLV-1 in Japan, multiple introductions of HTLV-1 in the past would be assumed. The end at the beginning…