Phylogeny: Principles & Molecular Approaches - Biology Presentation
advertisement

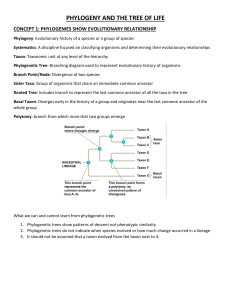
CHAPTER 8 PHYLOGENY: BASIC PRINCIPLES AND MODERN MOLECULAR APPROACHES GENERAL BIOLOGY 2 – 12 STEM KEY OBJECTIVES: •Describe species diversity procedures to establish evolutionary histories. •Explain how modern systematists use DNA sequences data to answer the perplexing concepts of speciation and evolution. • Classifications should be “natural”, meaning they reflect evolutionary relationships as closely as possible. • We do not, for example, place slime molds and eagles in the same family. • Systematics is a two-part endeavor. First, construct a hypothesis of evolutionary relationship among the organisms under study and second, plan a classification scheme that would reflect the hypothesized relationship. • After understanding the principles of taxonomy and systematics on the previous chapter, this chapter would tackle the procedures of constructing evolutionary histories that use classical methodologies and modern molecular techniques. • This will introduce the basics of phylogenetic construction which is commonly used in showing evolutionary relationships. CONSTRUCTING THE CLADOGRAM • Phylogenies can be represented as tree-like diagrams called phylogenetic or evolutionary tree showing how various taxa branched out from their ancestors and from each other. • Different graphic representations of phylogenies. • Nodes represent common ancestors of taxa (branches). • The two lineages branching from the same ancestor arose at the same geologicaal time. A lot of people assume that Homo sapiens is the “most highly evolved” or most recently evolved species; however, neither is true because of the following rules. RULE #1: The branches at every mode can be rotated. The branches do not infer any sort of order; they indicate only how recent are the common descendants. RULE #2: Two lineages branching from a single ancestral node are known as sister taxa. The common ancestry is the main basis of taxonomic groupings and not based on subjective perceptions of specialization. RULE #3: There is no such thing as a “most highly evolved species”. It means that all extant species descended from successful ancestors, and evolved to survive and reproduce, adapted to their specific environment. RULE #4: No extant or extinct taxon is considered ancestral to any other extant or extinct taxon. This should be remembered when one hears the incorrect statement “ humans evolved from monkeys” because monkeys did not. Humans and monkeys share a common ancestor. GROUPING TAXA BASED ON SHARED APOMORPHIES • Using apomorphies, taxa can be grouped in different ways: A. Hennig Tree/Argumentation - uses characters one at a time. B. Wagner Tree/Method - taxa are connected one at a time until all the taxa are included in the tree. C. Rooted vs. Unrooted Trees. Trees can be rooted or unrooted, scaled or unscaled, or a combination of both. Rooted tree - used when each of the nodes represents the most recent common ancestor of the taxa branching from it. It is directional, which means that a single common ancestor is present at the root and all taxa evolve or radiate from that single common ancestor. Unrooted tree - used when there is no hypothetical ancestor, outgroup, or directionality to the tree and only the putative evolutionary relationships of the taxa on the tree are present in the tree. D. Monophyly, Paraphyly, and Polyphyly. A phylogenetic tree is not constructed randomly. A systematist uses different characters to determinee recency of common descent. One of the tasks of a systematist is to convert the tree into the formal hierarchical Linnaean classification by giving a formal taxonomic name to groups that share a common descent. Such groups are called monophyletic taxa and are recognized because they share unique derived characters. •Since the classification system was creted due to the absence of a clear guideline, some biologists called lumpers focus on similarities in organisms thereby delimiting several species into a single genus. •Other biologists called splitters focus on the differences between the species, thus, dividing the species into several different genera. • Systematists can change any classification schemes that were presented before because of the following reasons based on Lipscomb: 1. New data - input from new technologies provides new information on the similarities and differences among taxa that leads to revision, lumping, or splitting a taxon. 2. New taxa - discovery of new species with new or conflicting characterss leads to the revision of classification. 3. Misinterpreted data - new monophyletic taxon is created when new studies revealed that the characters used to group a certain taxon are not unique or are convergent with other characters. NEW TRENDS: MOLECULAR PHYLOGENETICS • It has since been determined that evolution is actually a molecular process based on genetic information, encoded in DNA, RNA, and proteins. • One molecule undergoes diversification into many variations while one or more of those variants can be selected to be reproduced or amplified throughout a population over many generations. • Indeed, the onset of DNA technology helped systematists to establish relationships between species and unknown species using genetic structures. • Evolutionary events are shaped by homology and represented in phylogenetic trees as homologous relationships. • Homologous relationships can occur as: 1. paralogs - homologous sequences separated by gene duplication 2. orthologs - separated by a speciation event A molecular phylogenetic tree is derived from biomolecular sequence alignments of DNA, RNA, or amino acis, molecular markers, such as single nucleotide polymorphisms (SNPs) or restriction fragment length polymorphisms (RFLPs), morphology data, or information on gene order and content. • According to Dowell, the process of creating a molecular phylogenetic tree is as follows: 1. DNA Isolation. A sample is taken from the specimen to be analyzed. 2. Amplification of the DNA. The DNA that was isolated is then amplified using the polymerase chain reaction. 3. DNA Sequencing. Now is the right time to know the actual and precise orders of nucleotides (A,T,C, and G) within the DNA molecule. There were a lot of different sequencing methods that were utilized in the past decades. 4. Dataset assembly. The real work begins after getting the whole actual DNA sequence. 5. Sequence alignment. After you selected and retrieved data, multiple sequence alignment is done. 6. Phylogenetic Tree Building. To construct a phylogenetic tree, statistical methods are applied to determine the tree topology and calculate the branch lengths that best describe the phylogenetic relationships of the aligned sequences in a dataset.