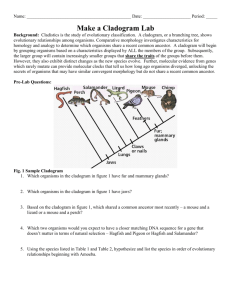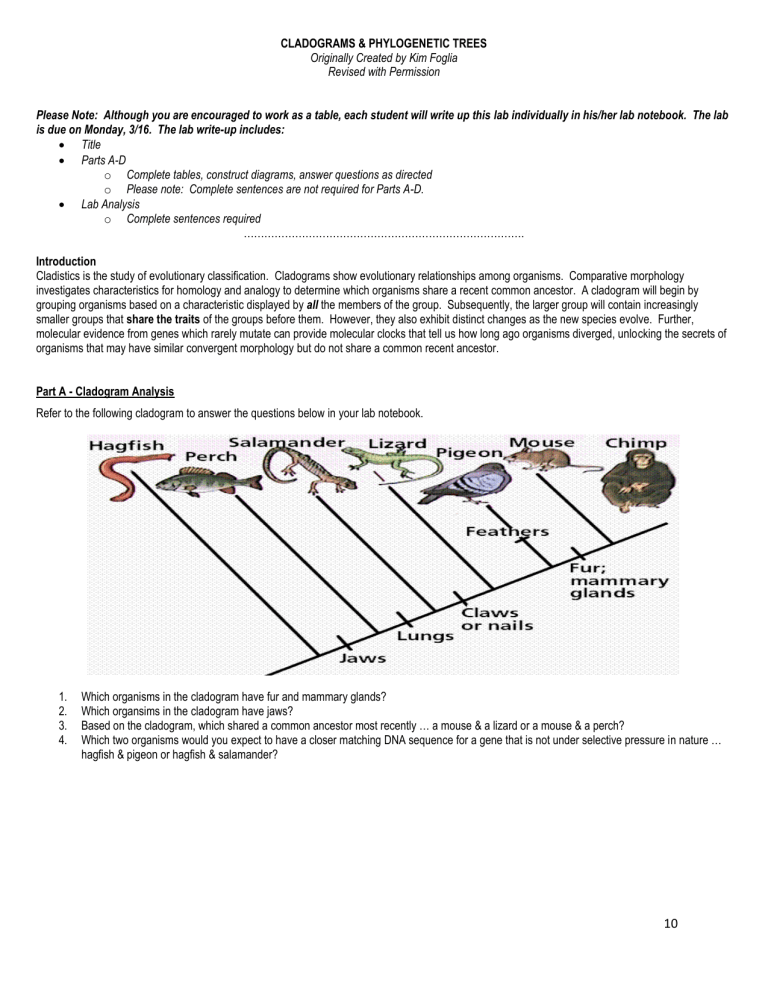
CLADOGRAMS & PHYLOGENETIC TREES Originally Created by Kim Foglia Revised with Permission Please Note: Although you are encouraged to work as a table, each student will write up this lab individually in his/her lab notebook. The lab is due on Monday, 3/16. The lab write-up includes: Title Parts A-D o Complete tables, construct diagrams, answer questions as directed o Please note: Complete sentences are not required for Parts A-D. Lab Analysis o Complete sentences required ………………………………………………………………………. Introduction Cladistics is the study of evolutionary classification. Cladograms show evolutionary relationships among organisms. Comparative morphology investigates characteristics for homology and analogy to determine which organisms share a recent common ancestor. A cladogram will begin by grouping organisms based on a characteristic displayed by all the members of the group. Subsequently, the larger group will contain increasingly smaller groups that share the traits of the groups before them. However, they also exhibit distinct changes as the new species evolve. Further, molecular evidence from genes which rarely mutate can provide molecular clocks that tell us how long ago organisms diverged, unlocking the secrets of organisms that may have similar convergent morphology but do not share a common recent ancestor. Part A - Cladogram Analysis Refer to the following cladogram to answer the questions below in your lab notebook. 1. 2. 3. 4. Which organisms in the cladogram have fur and mammary glands? Which organsims in the cladogram have jaws? Based on the cladogram, which shared a common ancestor most recently … a mouse & a lizard or a mouse & a perch? Which two organisms would you expect to have a closer matching DNA sequence for a gene that is not under selective pressure in nature … hagfish & pigeon or hagfish & salamander? 10 Part B – Cladogram Construction Based on Morphological Characteristics Examine the cladogram above illustrating the ancestry of these animals. Note that it reflects shared characteristics as time proceeds and how the different animals are all at the same time level (across the top) since they all live today. In addition to your vast array of knowledge, please use any classroom textbooks to determine the morphological characteristics of the organisms in the following table. Please Note: The universal common ancestor cell represents an out-group and does not possess any of the derived characteristics. Cut out the Part B Character table on the accompanying lab sheet and glue it into your Lab Notebook. Use the Hillis textbook provided, along with the cladogram on the previous page, to complete the Character Table. Mark the column with an X if the trait listed is essentially a characteristic of all organisms in the group described. For every characteristic the organism possesses, put an X in that box. The table is partially completed for you. Only 2 organisms listed share the same number of differences. Character Table Use this information to construct a cladogram in your Lab Notebook using the diagram below as a guide. Properly show all organisms and shared morphological characteristics. 10 Part C – Cladogram Construction Based on Molecular Evidence Cytochrome c is a protein located in the mitochondria of cells involved with cellular respiration. Cut out the table provided and glue it into your Lab Notebook. Compare each organism’s Cytochrome c DNA sequences with the ancestor cell and each other. o Use a ruler or straight edge to align the DNA nucleotides from every organism listed. o Circle or highlight the differences (mutations) present in the Cytochrome c DNA sequences from ancestor cell. o Determine the total number of mutations in each organism’s sequence as compared to the ancestor cell. Use the data to construct a cladogram in your Lab Notebook. Cytochrome c DNA Sequence Data Organism DNA Sequence # of mutations Ancestor Cell Amoeba ATTAGCGACCAGTATATCCTACAATCCGTCTACTTCATT 0 ATTAGCGACCAGTTTATCCTACAATCCCGTCTACTTCAT 11 Kangaroo CTAATCCCCCCGTTTATCCTACTTTCCCATCTACTAAGT Earthworm CTTATCGACCCGTTTATCCTACATTCCCGTCTACTTCGT Cat TTAATCCCCCCGTTTATCCTACTTTCCCATCTACTAAGT Shark CTTATCCCCCCGTTTATCCTACTTTCCCGTCTACTTCGT Dolphin CTAATCCCCCCGTTTATCCTACTTTCCCATGTAGTAAGT Lizard CTAATCCCCCCGTT TATCCTACTTTCCCGTCTACTTCGT Sponge ATTATCGACCAGTTTATCCTACATTCCCGTCTACTTCGT Part D – Phylogenetic Trees A phylogenetic tree is a tree showing the evolutionary interrelationships among various species that are believed to have a common ancestor. A phylogenetic tree is a form of a cladogram. In a phylogenetic tree, each node with descendents represents the most recent common ancestor of the descendents, and edge lengths correspond to time estimates. 10 Refer to the table below showing amino acid differences in the Cytochrome c protein of eight different species.. Construct a phylogenetic tree for the eight species listed above based on this information. Lab Analysis – Please answer in complete sentences. 1. Discuss how the cladogram constructed in Part B compared with the cladogram constructed in Part C. 2. Compare and contrast convergent evolution with divergent evolution. 3. Which two organisms show convergent evolution in the cladogram from Part B? Why might these two organisms have similar morphology despite not sharing a recent common ancestor? 4. The evolution of a species is dependent on changes in the genome of the species. Identify two mechanisms of genetic change and explain how each affects genetic variation. 5. The Cytochrome c gene is described as highly conserved. Discuss what this means in physiological and evolutionary terms. 6. When amino acid changes occur, what are two possible explanations for why they are survivable? 10