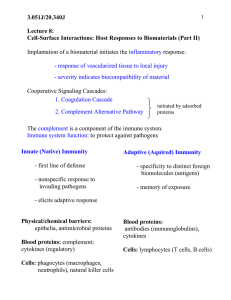
Complement System
advertisement

Complement Piet Gros Crystal and Structural Chemistry Bijvoet Center, Utrecht University The Netherlands Images, movie, texts: h@p://www.crystal.chem.uu.nl/~gros/complement.php Effects from complement • Immune defense – Gram-­‐negaJve bacteria • GeneJc changes in complement proteins – Kidneys: atypical hemolyJc uremic syndrome (aHUS) and dense-­‐ deposit disease (DDD) – Eyes: Age-­‐related macular degeneraJon (AMD) • Pathological condiJons affected by complement – Chronic inflammaJon – Acute trauma e.g. strokes and heart a@acks – CNS: schizophrenia & Alzheimer’s long-­‐term memory problems – PeriodonJJs Complement has a general role in Tissue Maintenance Host protection Complement proteins • ProteolyJc cascade – Network of large modular proteins – Complex formaJon, protease acJvaJon, proteolysis, … • RegulaJon – Rapid and localized response – IniJaJon, localizaJon, amplificaJon, inhibiJon and effector funcJons Ini6a6on Labelling and amplifica6on Effector Protec6on C1: C1q C1r C1s C4 C3 C5 Factor H MBL-­‐MASP2 C2 Factor B C6 C7 C8 C9 MCP Factor D CR2 DAF Properdin CR3, CR4 CR1 Ficolins-­‐MASP2 C4BP Proteins coloured by homology What are the molecular mechanisms underlying complement? IniJaJon Covalent labelling and amplificaJon Macrophage (1) Classical pathway (anJbodies) (2) LecJn pathway (mannans) (3) AlternaJve pathway (“Jck-­‐over”) B-­‐cell sJmulaJon phagocytosis C5 C4 C1 C3 FB & FD C2 FI C3b Ab Effector funcJons B cell inflammatory responses C6 C7 C5b C8 lysis C9 C3(H2O) (1) (2) convertase bacteria, fungi, virus, … altered host cell, Jssue debris (3) convertase host protecJon Complement regulators opsonizaJon MAC What are the molecular mechanisms underlying complement? AnJbody mediated complement acJvaJon Membrane A@ack Complex Diebolder, Beurskens et al. Science 2014 AlternaJve pathway: C3, FB &FD C3 Host protecJon: regulators & FI FB & FD FI C3b C3(H2O) convertase Sharp et al. Cell Reports 2016 What are the molecular mechanisms underlying complement? β C3: 1 FB: α Ba CCP1 CCP2 CCP3 aL VWA 1,641 * Bb SP 739 Arg-­‐234 – Lys-­‐235 FD: FI: FH (CCP1-­‐20) CR1 (1-­‐30) MCP (1-­‐4) DAF (1-­‐4) Regulators: AlternaJve pathway: C3, FB & FD C3 FB & FD Host protecJon: regulators & FI FI C3b C3(H2O) convertase Alterna6ve Pathway • Tick-­‐over acJvated (no specificity!) • AmplificaJon by proteases factor B and D • Yields opsonizaJon with (covalently a@ached) C3b • How is it iniJated? • How is it amplified? • How is it controlled/stopped? • Data from structural, biophysical and mutagenesis studies Proteoly6c cleavages determine the state of C3 C3 1 β α 1,641 * • Dormant Cleavage by C3 convertase C3a C3b * Factor I • AmplificaJon • Phagocytosis • Cell lysis C3f iC3b * Factor I • Phagocytosis • B cell sJmulaJon • InacJve C3c C3dg * • B cell sJmulaJon Structure of C3 MG1-­‐8 Bert Janssen ANA (C3a) Bert Janssen et al. Nature 2005 CUB LNK C345c TED (C3d) In coll. with Moh Daha (Leiden), Bo and KrisJna Nilsson (Uppsala) Evolu6onary implica6ons CUBgf 743 578 MG6βα 912 1330 806 535 β chain 962 1269 α chain ANA LNK (C3a) TED (C3d) CUB MG1 HypotheJcal Ancestor: MG2 MG3 MG4 β chain MG5 MG6 MG7 MG8 C345C (RBD) (NTR) α chain Reac6ve thioester required for surface aUachment is protected MG8 Tyr1460 Met1378 Tyr1425 CUB Phe1047 MG8 TED TED MG2 Gln991 Cys988 Hydrophobic/aromaJc pocket protects the thioester against hydroxyl and amino nucleophiles C3 ? Ac6va6on by proteolysis C3b Thioester in C3 Acylimidazole Intermediate in C3b and C3d C3d: Nagar et al. Science 1998 Law & Dodds, Prot. Sc. (1997) In coll. with John Lambris (USA) Bert Janssen et al. Nature 2006 Structures of C3 and its fragments C3a C3c C3b C3 iC3b C3dg C3a: C3d: C4d: C3 & C3c: C3b: C3b: bovine C3: Insect TEP: C5: C4: Huber et al. Hoppe-­‐Seyler 1980 Nagar et al. Science 1998 Van den Elsen et al. JMB 2002 Janssen et al. Nature 2005 Janssen et al. Nature 2006 Wiesmann et al. Nature 2006 Fredslund et al. JMB 2006 Baxter et al. PNAS 2007 Fredslund et al. Nature Imm 2008 Kidmose et al. PNAS 2012 How is the complement response amplified? Bb C3 B Mg2+ fD 90 s C3b [host protecJon] • How is the C3 convertase formed by factor B and D ? -­‐ formaJon of proconvertase C3bB -­‐ acJvaJon of D -­‐ cleavage of B • How is C3 acJvated by the C3 convertase ? • … why is this a mulJ-­‐step process? Assembly driven by proteolyis and large conforma6onal changes 84° 134° Mg2+ Janssen et al. EMBO J 2009 Rooijakkers, Wu et al. Nature Imm 2009 Forneris et al. Science 2010 SEE WEBSITE: http://www.crystal.chem.uu.nl/~gros/movies.php Alternative Pathway: APmovie.short.notext.nomusic.speed30fps.v1.0.avi (125 MB) Modelled Modelled C3b-­‐Factor aFactor Factor C3bBD wer C3bB C3b C3 a(wer C3bBb-­‐SCIN) (D 2a73) B (H (2i07) ( 2xwj) 2xwb) (1-­‐4) (C2xw9) 2ok5) VF-­‐B 34(4.3 .0 2wii) 3.0 (213hrz) .5 ÅÅ .3 .2 Å, 2, Å R , Å R (Å22win) , Rw, w, .7 R= R = 2Rw= .2 0wÅ 0= = 0.23, = .27, , Å 0.23, 0R 30.19, , .20, .9 .15, R= RRwÅ Rff0= = Rf, R .22, = R0fR0= 0.18, .29 = .32 0.28 = 00.24 R.24 .18 0f= R .25, = .25 0R.23 w w w ff= w f0 f= 0.27 Rooijakkers, Wu Milder Forneris Janssen Forneris eJanssen t W al. u eN et ature t eaeea t l. t t l. a N aal. N l. ature l. IEmmunology ature SN SMBO cience cience ature SIMB mmunology J. 222005 006 010 009 2010 007 2010 2009 Convertase dissociates irreversibly 84° 134° Why irreversible dissocia6on? C3b binding site SPR data C3b-B: VWA kon = 2.9 x 104 M-1s-1 (fast) koff = 0.1 s-1 (fast) kon = 1.1 x 10-2 M-1s-1 (slow) koff = 2.6 x 10-3 s-1 (slow) SP Slow kinetics Mg2+ dependent C3b:Ba ~4,000 Å2 “engineered” Bb (free) Bb (free) Bb (complex) C3b:VWA ~1,000 Å2 Mg2+ Not explained by structural changes. In coll. with Daniel Ricklin and John Lambris (UPenn, USA) Ba needed for loading of FB, Bb slow and irreversible release Assembly yields ‘6med’ convertase • Pro-­‐pepJde Ba needed for loading of FB (fast on rate) • Protease fragment releases slowly and irreversibly, due to on/off rates at Mg2+ (MIDAS) binding site Alterna6ve Pathway: amplifica6on • • • • • • Cleavage of C3 to C3b C3 convertase formaJon by FB and FD Ba needed for loading C3bBb enzyme complex C3bBb half-­‐life Jme ~ 1 min, dissociates irreversibly Build up by C3b-­‐B-­‐D needed to localize (through C3b), amplify (through C3bBb) and stop (through slow on/off kineJcs of C3b-­‐Bb) • Homology of alternaJve vs. classical and lecJn pathways: – AP: C3 , B, D – CP: C4, C2, C1qr2s2 and LP: C4, C2 , MBL/Ficolin MASP2 Host protec6on by regulators Bb [inflammatory responses] C3 C3bBb 90s fI [recogniJon] Regulators Regulators use different mechanisms: • • [cell lysis] [phagocytosis] [B-­‐cell sJmulaJon] Decay-­‐acceleraJng acJvity (DAA): Convertase dissociaJon Blocking pro-­‐convertase formaJon Cofactor acJvity (CA): AssisJng factor I in degrading C3b iC3b C3d Host protec6on by regulators Bb [inflammatory responses] C3 C3bBb [cell lysis] [phagocytosis] [B-­‐cell sJmulaJon] 90s fI [recogniJon] iC3b C3d Regulators FH CR1 CCP/SCR/Sushi soluble DAA/CA DAA/CA CA/DAA DAF DAA MCP CA SPICE VCP CA/DAA CA/DAA CA/DAA Small-­‐pox virus Vaccinia virus human Structure of C3b-­‐FH (1-­‐4) α’NT Hocking et al. JBC 2008: CCP1-­‐2 (NMR) CCP2-­‐3 (NMR) CCP1 CCP1-­‐4 (X-­‐ray in complex with C3b) CCP3 CCP2 CCP4 • • • • TED 100 Å long, disconJnuous interface 4,800 Å2 buried surface area KD = 7 µM MAb blocking studies confirm interacJon sites Jin Wu et al. Nature Immunology 2009 in coll. with Ricklin & Lambris (UPenn) Implica6ons for decay accelera6on C3b-­‐FH(1-­‐4) C3bBb DAF CCP2-­‐4 superimposed DAA: CCP1-­‐2 DAF mutagenesis data: Ku@ner-­‐Kondo et al. JBC 2007 DAF structure: Lukacik et al. PNAS 2004 Jin Wu et al., Nature Immunology 2009 Implica6ons for DAA and CA C3b-­‐FH(1-­‐4) C3bBb CA: CCP1-­‐3 Factor I Roversi et al. PNAS 2011 DAA: CCP1-­‐2 Jin Wu et al., Nature Immunology 2009 Do all regulators bind C3b in the same way? InteracJons with α’-­‐NT loop are important for FH and CR1, but not MCP. Lambris et al. JIM (1996), Oran et al. JBC (1999) Mapping MCP mutants on C3bFH(1-­‐4) α’NT CCP1 CCP2 CCP3 CCP4 C3bFH(1-­‐4) MCP mutant C3b binding Wild type 100% K193A 37% R195A 50% F196A 35% F208A 30% K210A 68% K211A 56% K212A 32% MCP mutants data: Liszewski et al. JBC (2000) • The mutagenesis data suggest that MCP binds differently. Structure of C3b-­‐MCP at 2.4-­‐Å resolu6on MCP was provided by Sfyroera and Lambris (UPenn) MCP C3b binding construct Wild type 100% ΔCCP1 No effect MCP ΔCCP2 ΔCCP3 ΔCCP4 MCP constructs data: Adams et al. JBC (1991) CCP4 120° C3b MCP mutants data: Liszewski et al. JBC (2000) • No density for CCP1 and CCP2 • CCP4 differs 120° wrt FH CCP4 Wu, Forneris, Xue et al. in preparaJon C3b in complex with regulators Res.: 2.4 Å 2.7 Å 2.7 Å 3.6 Å 3.4 Å CA CA/daa CA/DAA CA/daa DAA C3b-­‐MCP KD = 0.35 µM C3b-­‐SPICE 1 µM C3b-­‐FH(1-­‐4) 7 µM C3b-­‐CR1(15-­‐17) 2 µM C3b-­‐DAF(2-­‐4) 14 µM In coll. with Daniel Ricklin, John Lambris (USA) and John Atkinson (USA) Xue, Forneris, Wu et al., EMBO J 2016 C3b in complex with regulators Res.: 2.4 Å 2.7 Å 2.7 Å 3.6 Å 3.4 Å CA CA/daa CA/DAA CA/daa DAA C3b-­‐MCP KD = 0.35 µM C3b-­‐SPICE 1 µM C3b-­‐FH(1-­‐4) 7 µM C3b-­‐CR1(15-­‐17) 2 µM C3b-­‐DAF(2-­‐4) 14 µM In coll. with Daniel Ricklin, John Lambris (USA) and John Atkinson (USA) Disease related mutants of C3b and FH(1-­‐4/19-­‐20) H1442D acJvaJon routes C3a C3 FD R570W/Q C3 convertase (C3bBb) P240L K204Δ D1093N A1072V Q1139K W1157L L1189R D1119G Diseases: aHUS, MPGN-­‐II, AMD iC3b FB Ba C3b FH:C3b FI FH19-­‐20 Gros, Nature SMB 2011 T1184R R1210A R1215Q C3d-­‐FH(19-­‐20): Kajander et al. PNAS 2011 Morgan et al. Nature SMB 2011 C3d R109L R60G V44I R35H (either specific or non-­‐specific) Structural basis of complement IniJaJon Covalent labelling and amplificaJon Macrophage Classical pathway (anJbodies) LecJn pathway (mannans) AlternaJve pathway (“Jck-­‐over”) C1 MBL C4+C2 B-­‐cell sJmulaJon CR2-­‐C3d phagocytosis C3 C3a C3b inflammatory responses C5 Factor B and factor D Ab C3(H2O) Effector funcJons B cell Factor I iC3b C6 C7 C5b C8 lysis C9 C3d C3b opsonizaJon host protecJon Immune evasion: SCIN, E}, Ecb, FHbp, … Complement regulators: FH, MCP, DAF, CR1, CRIg, SPICE MAC CP on vesicles by cryo-­‐electron tomography Christoph Diebolder DNP-­‐liposomes + anJ-­‐DNP Ab + NHS height: ~25 nm DNP-­‐liposomes + anJ-­‐DNP Ab height: ~11 nm In coll. with Roman Koning and Bram Koster (LUMC) EM tomography of C1 binding DNP-­‐liposomes + anJ-­‐DNP Ab + purified C1 or C4 depleted serum Height of Ab-­‐C1 complex: ~35 nm Cryo-­‐ET subtomogram average 107 parJcles ResoluJon ~6 nm 10 nm IgG hexamer fits lower pladorm 6x Fc ring, 6x 1 Fab equatorial and 1 Fab axial 180° Fab’s up/down – Fab’s all down PDB: 1HZH: Saphire, Science (2001) 6-­‐fold symmetrized map Fieng C1 into the complex Unsymmetrized Symmetrized Consistent with extensive literature -­‐ C1 data, SAXS, EM, Xray fragments -­‐ Fc-­‐C1q mutagenesis Christoph Diebolder, Frank Beurskens et al. Science 2014 Hexameriza6on can be enhanced and is general applicable Bispecific IgG induce CDC Classical Pathway: specific ini6a6on • Geometry determines propensity of hexamer formaJon – AnJgen density and fluidity – PosiJon of epitope on anJgen – Height of epitope from membrane (2 reasonings) • AcJvaJon of C1r2s2 sJll quite unclear • DeposiJon kineJcs/spaJal topology sJll quite unclear • Homology of classical and lecJn pathways: – CP: C1qr2s2 , C4, C2 – LP: MBL/Ficolin MASP2, C4, C2 Structural basis of complement IniJaJon Covalent labelling and amplificaJon Macrophage Classical pathway (anJbodies) LecJn pathway (mannans) AlternaJve pathway (“Jck-­‐over”) C1 MBL C4+C2 B-­‐cell sJmulaJon CR2-­‐C3d phagocytosis C3 C3a C3b inflammatory responses C5 Factor B and factor D Ab C3(H2O) Effector funcJons B cell Factor I iC3b C6 C7 C5b C8 lysis C9 C3d C3b opsonizaJon host protecJon Immune evasion: SCIN, E}, Ecb, FHbp, … Complement regulators: FH, MCP, DAF, CR1, CRIg, SPICE MAC Membrane attack complex [inflammatory responses] C5 C5a C5b C3b C7 C6 C5b-­‐7 C5 convertase MACPF C6 C7 γ C8 β C9 Perforin C9 C9 ~12-­‐20 copies of C9 C9 C5b6 α C8 C5b-­‐8 MACPF resembles bacterial cholesterol-dependent cytolysins N C8γ C Human C8α-­‐MACPF Bacterial CDC Michael Hadders et al. Science 2007 Sodetz, JMB 2008 Rosado et al. Science 2007 Poleknina et al. PNAS 2005 Shephard et al., Biochemistry (1998) Shatursky et al., Cell (1999) First step in MAC formation C5b C6 C5b6 C7 C8 C9 C9 C9 C5b6 C5b C5b-7 C5b-8 MACPF C6 Tschopp et al., PNAS (1982) C8: Lovelace et al. C6: Aleshin et al. JBC 2011 C8α MACPF: Michael Hadders et al. Science 2007 C5b6: Michael Hadders, Doryen Bubeck et al. Cell Reports 2012 C5b6: Aleshin et al. JBC 2012 Change in C5b captured by C6 C3 to C3b C5 to C5b6 C5: Fredslund et al. Nature Imm. 2008 C5b C6 C5b6 C5b7 C5b8 C7 C5b-7 C8 C5b-8 C9 C9 C9 MAC pore formation MAC pores Sub-tomogram averaging In situ MAC has seam above membrane C5b C6MACPF C6-CCP/ FIM C7 C8βα C8γ C9 Volta phase plate, Krios Titan, Gatan K2 73 tomograms, 986 subvolumes, EMAN2, 21 Å resoluJon Thom Sharp et al. Cell Reports 2016 Cone-shaped pore with seam, aggregated pores We observe diverse oligomers, but no hemi-pores. Recogni6on and regula6on in Complement • RecogniJon/iniJaJon – CP: C1 binding to anJbodies and directly to “danger” signals – LP: glycan recogniJon – AP: low level of Jck over • Complex formaJon and conformaJonal changes determine regulaJon in the complement system – – – – – Sequence of proteolyJc acJvaJons AmplificaJon within ~ 1min by C3 convertase C3b opsonizaJon Host protecJon by regulators Pore formaJon by MAC


![Anti-C3d antibody [7C10] ab17453 Product datasheet 1 Abreviews 2 Images](http://s2.studylib.net/store/data/012480916_1-f57c10e193fe8f35889da837e825ca42-300x300.png)