Priniciples of transplantation February 19 2008
advertisement

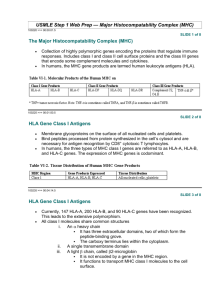
Priniciples of transplantation February 19 2008 Goals and objectives - Principal components - Role of HLA in immunogenic response - Understand the 3 signal pathway of T cell activation and its clinical significance Classification of grafts * Autologous grafts Grafts transplanted from one part of the body to another in the same individual * Syngeneic grafts (Isografts) Grafts transplanted between two genetically identical individuals of the same species Allogeneic grafts (Allografts) Grafts transplanted between two genetically different individuals of the same species Xenogeneic grafts (Xenografts) Grafts transplanted between individuals of different species 2006-7year Immunology 4 IMMUNE RESPONSES TO TRANSPLANTED TISSUES * Transplant rejection caused by genetic differences between donor and recipient * HLA and blood group antigens * Alloantigens * Antigens which vary between members of same species * Alloreaction * Immune response to an alloantigen * Alloreactions in transplantation * Host-versus-graft (transplant rejection) * Graft-versus-host Figure 12-11 Effectors of rejection * * * * Major players: T Cells B cells Antigen presenting cells MHC (Most important) T cells * Arise in thymus from bone marrow derived precursors * Each T-Cell has unique T Cell receptor (Clone) * Selection Positive Negative * Subtypes CD 4 T cells – Antigen specific immune response CD8 T cells - Precursors of CTL – Class I MHC B cells * * * * Arise and mature in bone marrow Negative selection Express BCRs on their surface When BCR is stimulated the B cell secrete antibodies of same specificity as their BCRs Antigen presenting cells * Most important * Activate T cells * Endocytose antigen and display it on MHC molecules * T cells recognize and interact with antigen MHC to become activated MHC complex * Encode molecules crucial to the initiation and propagation of immune response * The HLA complex on chromosome 6 contains over 200 genes, more than 40 of which encode leukocyte antigens * The HLA genes that are involved in the immune response fall into two classes, I and II, which are structurally and functionally different Location and Organization of the HLA Complex on Chromosome 6 Klein J and Sato A. N Engl J Med 2000;343:702-709 Types of MHC * There are three classes of MHC molecules. * Class I- encodes glycoproteins expressed on the surface of nearly all nucleated cell; the major function of the class I gene is presentation of peptide antigens to cytotoxic T-cells * Class II- encodes glycoproteins expressed primarily on antigen-presenting cells, examples: macrophages, dendritic cells and B-cells, where they present processed antigenic peptides to T helper cells. * Class III- encodes various secreted proteins that have immune function including components of the complement system; C2,C4, Factor B, &TNF, and molecules involved in inflammation. Nucleated cells Class I MHC RBCs Class II MHC APCs Function of MHC * The function of both class I and class II molecules is the presentation of short, pathogen-derived peptides to T cells, a process that initiates the adaptive immune response * Class I - Sample cytosolic proteins and detect foreign proteins that would indicate an intracellular pathogen such as virus or intracellular bacteria * Recognised by CD 8 T cells and provide a surviellance mechanism to target infected cells for destruction Antigen Processing and Presentation Biological functions of Class I and Class II molecules * http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/H/HLA.html#cd8 Class I * Present peptides derived from endogenously synthesized proteins * Responding T cells express CD8+ * Class II system is designated to sample extracellular proteins by extracellular proteins by specialized APC’s * Class II are recognized by CD 4 helper T cells and allow for the generation of immune response to invading pathogens Antigen Processing and Presentation Biological functions of Class I and Class II molecules * Class II * Present peptides derived from exogenously synthesized proteins * Responding T cells express CD4+ http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/H/HLA.html#class_II Class I * The class I genes code for the polypeptide chain of the class I molecule; the ß chain of the class I molecule is encoded by a gene on chromosome 15, the beta2microglobulin gene. * There are some 20 class I genes in the HLA region; three of these, HLA-A, B, and C, the so-called classic, or class Ia genes, are the main actors in the immunologic theater Structure of Class I MHC * Two polypeptide chains, a long α chain and a short β (β2 microglobulin) * Four regions * Cytoplasmic region containing sites for phosporylation and binding to cytoskeletal elements * Transmembrane region containing hydrophobic amino acids Structure of Class I MHC * * Four regions * A highly conserved α3 domain to which CD8 binds * A highly polymorphic peptide binding region formed from the α1 and α2 domains Β2-microglobulin helps stabilize the conformation Structure of HLA Class I and Class II Molecules Klein J and Sato A. N Engl J Med 2000;343:702-709 Class II * The class II genes code for the alpha and ß polypeptide chains of the class II molecules . * The designation of their loci on chromosome 6 consists of three letters: the first (D) indicates the class, the second (M, O, P, Q, or R) the family, and the third (A or B) the chain ( or ß, respectively). * HLA-DRB, for example, stands for class II genes of the R family coding for the ß chains. Structure of Class II MHC * * Two polypeptide chains,α and β, of roughly equal length Four regions * Cytoplasmic region containing sites for phosporylation and binding to cytoskeletal elements Structure of Class II MHC * Four regions * Transmembrane region containing hydrophobic amino acids * A highly conserved α2 and a highly conserved β2 domains to which CD4 binds * A highly polymorphic peptide binding region formed from the α1 and β1 domains Structure of HLA Class I and Class II Molecules Klein J and Sato A. N Engl J Med 2000;343:702-709 Important aspects of MHC * * * Normally, the proteins that undergo recycling are the organism's own, but in infected cells, proteins originating from the pathogen are also routed into the processing pathways. With the exception of jawed vertebrates, no organisms appear to make a distinction between peptides derived from their own (self) proteins and those derived from foreign (nonself) proteins. Jawed vertebrates, by contrast, use the peptides derived from foreign (usually microbial) proteins to mark infected cells for destruction Important aspects of MHC * * Protein processing and loading of peptides onto class I molecules are taking place all the time in most cells. There is always plenty of material to feed the processing machinery, because worn-out, damaged, and misfolded proteins are continuously being degraded and replaced by new ones. By contrast, the processing of exogenous proteins and the loading of peptides onto class II molecules are normally restricted to B cells, macrophages, and dendritic cells, which are very efficient in taking up material by endocytosis or phagocytosis. Important aspects of MHC * * * * The consequence of protein processing is that the surfaces of cells become adorned with peptide-laden HLA molecules, amounting on a per cell basis to roughly 100,000 to 300,000 class I or class II products of each of the highly expressed HLA loci. Since each HLA molecule has one peptide bound to it, each uninfected cell displays hundreds of thousands of self peptides on its surface. Each cell thus displays a heterogeneous collection of peptides, and the surface of a cell resembles rows of well-stocked stalls at a bazaar, with bargain hunters scrutinizing the wares. But if, in this metaphor, the vendors are the HLA molecules and the peptides the goods, who are the potential buyers? They are a group of lymphocytes reared in the thymus and then turned loose to roam the body — the T cells. Functions and Characteristics of HLA * HLA’s are cell-surface proteins involved in the recognition of self and non-self by the immune system * HLA’s present foreign antigens to the immune system – resistance to viral and bacterial pathogens * HLA’s are codominantly expressed * Highly polymorphic and polygenic HLA genes are co dominant: A protein from each parental gene is expresed on cellsurfaces Polymorphism and polygeny * MHC genes are polymorphic: that is, there are large numbers of alleles for each gene * MHC genes are polygenic: that is, there are a number of different MHC genes. Crossing over results in new haplotypes Structure of Class I MHC Variability map of Class 1 MHC α Chain Class I polymorphism Locus HLA - A Number of alleles (allotypes) 451 HLA - B 782 HLA - C 238 There are also HLA - E, HLA - F and HLA - G Relatively few alleles Structure of Class II MHC Variability map of Class2 MHC β Chain Class II polymorphism Locus Number of alleles (allotypes) HLA - DPA HLA - DPB 147 HLA - DQA HLA - DQB 105 HLA - DRA HLA - DRB1 HLA – DRB3 HLA – DRB4 HLA – DRB5 525 There are also HLA - DM and HLA - DO Relatively few alleles Why polymorphic? * Multiple alleles of HLA in a population increases the likelihood that the population will survive a pathogen threat * Unfortunately, it also cause histoincompatibility in organ and tissue transplants Important Aspects of MHC * Primary HLA products that contribute to rejection are the most polymorphic including HLA- A, - B and DR * Efforts are made to match HLA-A, - B and DR genes and proteins in kidney transplantation HLA profiles * Tissue typing * Cross matching test * Panel reactive antibodies Tissue typing * Helps to identify two alleles at each of the three loci * One allele from mother and one from father * Mother/Father: 25% chance of full match * One Sibling: 25 % chance of full match * Two Siblings: 44 % chance of full match * HLA matching 3 year graft surivival 93 and 85% for HLA matched and mismatched live donors In cadaveric grafts 82 and 76% Most benefit with zero mismatches Cross matching test * Serum of potential recipient is incubated with cells from possible donor * If recipient has antidonor antibodies there is a strong likelihood that recipient would destroy transplant by antibody mediated rejection Panel reactive antibodies * Anti HLA antibodies in the serum of a person can be assessed as PRAs * Testing the serum of the patient against a panel of cells or antigens prepared from many different donors using cytotoxicity or flow cytometry * Results are expressed as percentage of positive donors Response to antigenic stimulus T cell activation * Activation of T cell is a crucial step in generation of immune response to specific antigens * Naive T cells restricted to SLOs * They interact with DCs that have migrated from the periphery in response to infection or injury * Once naïve T cells encountered their cognate antigen presented on mature DCs, they become activated. * Following activation CD 4 T cells help B cells to convert to plasma cells * Plasma cells produce antibodies Generation of T cell effector function * Alloreactive T cells can be found in both naive and memory T cells * Naive T cells may be triggered by donor or recipient APCs to proliferate and develop effector functions in SLOs * Memory cells can be activated in the same manner or by recognizing cells in allograft directly * Reactions mediated by naïve T cells take longer to develop than those mediated by memory T cells Structure of the T cell Receptor * Heterodimer with one α and one β chain of roughly equal length * A short cytoplamic tail not capable of transducing an activation signal * A transmembrane region with hydrophobic amino acids Structure of the T cell Receptor * Both α and β chains have a variable (V) and constant (C) region * V regions of the α and β chains contain hypervariable regions that determine the specificity for antigen Structure of the T cell Receptor * Each T cell bears TCRs of only one specificity (allelic exclusion) TCR and CD3 Complex * TCR is closely associated with a group of proteins collectively called the CD3 complex * * * * γ chain δ chain 2 ε chains 2 ξ chains * CD3 proteins are invariant Role of CD3 Complex * CD3 complex necessary for cell surface expression of TCR during T cell development * CD3 complex transduces signals to the interior of the cells following interaction of Ag with the TCR The “Immunological Synapse” * The interaction between the TCR and MHC molecules are not strong * Accessory molecules stabilize the interaction * CD4/Class II MHC or CD8/Class I MHC * CD2/LFA-3 * LFA-1/ICAM-1 The “Immunological Synapse” * Specificity for antigen resides solely in the TCR * The accessory molecules are invariant * Expression is increased in response to cytokines The “Immunological Synapse” * Engagement of TCR and Ag/MHC is one signal needed for activation of T cells * Second signal comes from costimulatory molecules * CD28 on T cells interacting with B7-1 (CD80) or B7-2 (CD86) * Others * Costimulatory molecules are invariant * “Immunological synapse” Costimulation is Necessary for T Cell Activation * Engagement of TCR and Ag/MHC in the absence of costimulation can lead to anergy * Engagement of costimulatory molecules in the absenece of TCR engagement results in no response * Activation only occurs when both TCR and costimulatory molecules are engaged with their respective ligands * Downregulation occurs if CTLA-4 interacts with B7 * CTLA-4 send inhibitory signal Clonal expansion * Signals 1 and 2 activate calcium calcineurin pathway, MAP kinase pathway and NF-Kb pathway * These pathways activate transcription factors that trigger the expansion of many new molecules including IL-2, CD 154 and CD 25. * IL-2 and other cytokines activate the TOR pathway to provide signal 3, the trigger for cell proliferation * A subset of activated helper T cells migrate to the B region of lymph nodes located in the cortex and help to differentiate B cells while the remainder of the effector T cells leave the lymph node and proceeds the inflamed site * The activated T cells rapidly accumulate in the interstitium of the allograft as the response mounts in the first few days. * CD4 T cells are cytokine secreting cells that express IL-2 and alter a variety of cytokines * CD4 cells help B cells to enhance their antibody production through CD40 ligand * Alloantibody produced during rejection is mainly IG g and primarily participates in the destruction of vascular endothelium of the graft * CD 8 T cells participate in rejection through DTH or cytotoxicty Key Steps in T cell Activation * APC must process and present peptides to T cells * T cells must receive a costimulatory signal * Usually from CD28/B7 * Accessory adhesion molecules help to stabilize binding of T cell and APC * CD4/MHC-class II or CD8/MHC class I * LFA-1/ICAM-1 * CD2/LFA-3 * Signal from cell surface is transmitted to nucleus * Second messengers * Cytokines produced to help drive cell division * IL-2 and others Rejection 1. Hyperacute rejection * Occurrence time * Occurs within minutes to hours after host blood vessels are anastomosed to graft vessels * Pathology * Thrombotic occlusion of the graft vasculature * Ischemia, denaturation, necrosis ORIGINS OF ANTIBODIES TO HLA AND ABO ANTIGENS IN HYPERACUTE REJECTION * Pregnancy * Fetus is allograft in mothers body * During birth, fetal cells can stimulate maternal immune response * Blood transfusion * HLA typing not performed for routine transfusion * Leukocytes and platelets in whole blood * Transplantation * Persons with more than one transplant Figure 12-15 * Complement activation * Endothelial cell damage * Platelets activation * Thrombosis, vascular occlusion, ischemic damage 2006-7year Immunology 77


![Anti-HLA DQ3 antibody [2HB6] ab20174 Product datasheet Overview Product name](http://s2.studylib.net/store/data/012451218_1-1dc1c11f13a3c3487a61cb14278554ed-300x300.png)