PBG 650 Advanced Plant Breeding Module 9: Best Linear Unbiased Prediction – Purelines

PBG 650 Advanced Plant Breeding
Module 9: Best Linear Unbiased Prediction
– Purelines
– Single-crosses
Best Linear Unbiased Prediction (BLUP)
•
Allows comparison of material from different populations evaluated in different environments
•
Makes use of all performance data available for each genotype, and accounts for the fact that some genotypes have been more extensively tested than others
•
Makes use of information about relatives in pedigree breeding systems
•
Provides estimates of genetic variances from existing data in a breeding program without the use of mating designs
Bernardo, Chapt. 11
BLUP History
• Initially developed by C.R. Henderson in the 1940’s
•
Most extensively used in animal breeding
• Used in crop improvement since the 1990’s, particularly in forestry
•
BLUP is a general term that refers to two procedures
– true BLUP – the ‘P’ refers to prediction in random effects models (where there is a covariance structure)
– BLUE – the ‘E’ refers to estimation in fixed effect models
(no covariance structure)
B-L-U
• “Best” means having minimum variance
• “Linear” means that the predictions or estimates are linear functions of the observations
•
Unbiased
– expected value of estimates = their true value
– predictions have an expected value of zero
(because genetic effects have a mean of zero)
Regression in matrix notation
Linear model
Parameter estimates
Y = X
+ ε b = (X’X) -1 X’Y
Source
Regression
Residual
Total df p n-p n
SS b’X’Y
Y’Y - b’X’Y
Y’Y
MS
MS
R
MS
E
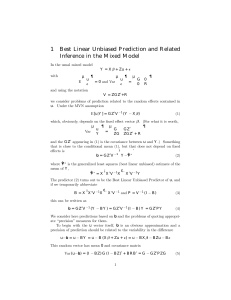
BLUP Mixed Model in Matrix Notation
Observations
Design matrices
Y = X
+ Zu + e
Residual errors
Fixed effects Random effects
•
Fixed effects are constants
– overall mean
– environmental effects (mean across trials)
•
Random effects have a covariance structure
– breeding values
– dominance deviations
– testcross effects
– general and specific combining ability effects
Classification for the purposes of
BLUP
BLUP for purelines – barley example
Environments
Set 1
Cultivar Grain Yield t/ha
18 Morex (1) 4.45
Set 1
Set 1
Set 2
Set 2
Set 2
18
18 Stander (4)
9
9
9
Robust (2)
Robust (2)
Excel (3)
Stander (4)
4.61
5.27
5.00
5.82
5.79
Parameters to be estimated
• means for two sets of environments – fixed effects
– we are interested in knowing effects of these particular sets of environments
• breeding values of four cultivars – random effects
– from the same breeding population
– there is a covariance structure (cultivars are related)
Bernardo, pg 269
Linear model for barley example
Y ij
=
+ t i
+ u j
+ e ij t i u j
= effect of i th set of environments
= effect of j th cultivar
In matrix notation: Y = X
+ Zu + e
4.45
1 0
4.61
1 0
5.27
= 1 0 b
1
5.00
0 1 b
2
5.82
0 1
5.79
0 1
1 0 0 0
0 1 0 0 u
1
+ 0 0 0 1 u
2
0 1 0 0 u
3
0 0 1 0 u
4
0 0 0 1 e
11 e
12
+ e
14 e
22 e
23 e
24
Weighted regression
Y = X
+ ε b = (X’X) -1 X’Y
Where ε ij
~N (0, σ 2 )
When ε ij
~N (0, R σ 2 )
Then b = (X’R -1 X) -1 X’R -1 Y
For the barley example
18 0 0 0 0 0
0 18 0 0 0 0
R
-1
= 0 0 18 0 0 0
0 0 0 9 0 0
0 0 0 0 9 0
0 0 0 0 0 9
Covariance structure of random effects
XY
Morex 1
Robust
Excel
Stander
Morex Robust Excel Stander
1/2
1
7/16
27/32
1
11/32
43/64
91/128
1
Remember
Cov
XY
r
2
A
2
D r = 2
XY
2 A u
A
2
2
1
1
2
7/8 11/16
27/16 43/32
7/8 27/16 2 91/64
11/16 43/32 91/64 2
A
2
Mixed Model Equations
βˆ uˆ
=
X’R -1 X
Z’R -1 X
X’R -1 Z
Z’R -1 Z + A -1 ( σ
ε
2 / σ
A
2 )
R σ 2
-1
X’R -1 Y
Z’R -1 Y
• each matrix is composed of submatrices
• the algebra is the same
Calculations in Excel
Results from BLUP
Original data
BLUP estimates
For fixed effects b
1 b
2
=
=
+ t
+ t
1
2
Environments
Set 1
Set 1
18
18
Cultivar
Morex
Robust
Grain Yield t/ha
4.45
4.61
Set 1
Set 2
Set 2
Set 2
18 Stander
9
9
9
Robust
Excel
Stander
5.27
5.00
5.82
5.79
1
2 u
1 u
2 u
3 u
4
Set 1
Set 2
Morex
Robust
Excel
Stander
4.82
5.41
-0.33
-0.17
0.18
0.36
Interpretation from BLUP
BLUP estimates
1
2 u
1 u
2 u
3 u
4
Set 1
Set 2
Morex
Robust
Excel
Stander
For a set of recombinant inbred lines from an F
2 cross of Excel x Stander
4.82
5.41
-0.33
-0.17
0.18
0.36
Predicted mean breeding value = ½(0.18+0.36) = 0.27
Shrinkage estimators
•
In the simplest case (all data balanced, the only fixed effect is the overall mean, inbreds unrelated)
BLUP (
i
)
h
2
Y i .
Y
..
•
If h 2 is high, BLUP values are close to the phenotypic values
•
If h 2 is low, BLUP values shrink towards the overall mean
•
For unrelated inbreds or families, ranking of genotypes is the same whether one uses BLUP or phenotypic values
Sampling error of BLUP
βˆ uˆ
=
X’R
Z’R
-1
-1
X
X
X’R -1 Z
Z’R -1 Z + A -1 ( σ
ε
2 / σ
A
2 )
-1 invert the matrix R σ 2
X’R -1 Y
Z’R -1 Y coefficient matrix
C
11
C
21
C
12
C
22 each element of the matrix is a matrix
•
Diagonal elements of the inverse of the coefficient matrix can be used to estimate sampling error of fixed and random effects
Sampling error of BLUP
βˆ
uˆ
=
C
11
C
21
C
12
C
22
X’R -1
Y
Z’R -1
Y
2
C
11
2
fixed effects
2
C
22
2
random effects
Estimation of Variance Components
(would really need a larger data set)
1.
Use your best guess for an initial value of σ
ε
2 / σ
A
2
2.
Solve for
ˆ and û
3.
Use current solutions to solve for σ
ε
2
σ
A
2 and then for
4.
Calculate a new σ
ε
2 / σ
A
2
5.
Repeat the process until estimates converge
BLUP for single-crosses
Performance of a single cross:
G
B73,Mo17
= GCA
B73
+ GCA
Mo17
+ SCA
B73,Mo17
BLUP Model
Y = X
+ Ug
1
+ Wg
2
+ Ss + e
•
Sets of environments are fixed effects
•
GCA and SCA are considered to be random effects
Example in Bernardo, pg 277 from Hallauer et al., 1996
Performance of maize single crosses
Set Entry Pedigree
1 SC-1 B73 x Mo17
1
1
SC-2
SC-3
H123
B84
x
x
Mo17
N197
2
2
SC-2 H123 x Mo17
SC-3 B84 x N197
Grain Yield t ha
-1
7.85
7.36
5.61
7.47
5.96
Iowa Stiff Stalk x Lancaster Sure Crop
7.85
7.36
5.61
7.47
5.96
1 0
1 0 b
1
= 1 0 b
2
0 1
0 1
1 0 0
0 0 1 g
B73
+ 0 1 0 g
B84
0 0 1 g
H123
0 1 0
1 0
1 0 g
Mo17
+ 0 1 g
N197
1 0
0 1
1 0 0
0 1 0 s
1
+ 0 0 1 s
2
0 1 0 s
3
0 0 1 e
11 e
12
+ e
13 e
22 e
23
Covariance of single crosses
SC-X is j x k SC-Y is j’ x k’
Cov
SC
jj '
2
GCA ( 1 )
kk '
2
GCA ( 2 )
jj '
kk '
2
SCA
B73, B84, H123
G
1
=
1
B73,B84
B73,B84
B73,H123
1
B84,H123
B73,H123
B84,H123
1 g1
MO17, N197
G
2
=
1
Mo17,N197
Mo17,N197
1
G 2
1 GCA(1) assuming no epistasis
g2
G
2
2 GCA(2)
Covariance of single crosses
SC-X is j x k SC-Y is j’ x k’
Cov
SC
jj '
2
GCA ( 1 )
kk '
2
GCA ( 2 )
jj '
kk '
2
SCA
SC-1= B73 x MO17 SC-2= H123 x MO17 SC-3= B84 x N197
1
S =
B73,H123
Mo17,Mo17
B73,B84
Mo17,N197
B73,H123
Mo17,Mo17
1
B73,B84
Mo17,N197
B84,H123
Mo17,N197
B84,H123
Mo17,N197
1 s
S
2
SCA
Solutions
X'R
-1
X X'R
-1
U
U'R
-1
X U'R
-1
U +
Q
1
W'R
-1
X W'R
-1
U
Z'R
-1
X Z'R
-1
U
X'R
-1
W
U'R
-1
W
W'R
-1
W +
Q
2
Z'R
-1
W
X'R
-1
Z
U'R
-1
Z
W'R
-1
Z
Z'R
-1
Z +
Q
S
-1
X
X'R
-1
Y
U'R
-1
Y
W'R
-1
Y
Z'R
-1
Y
G
1
1
G
2
1
Q
S
S
1
/
/
/
2
GCA(1)
2
GCA(2)
2
SCA
b
1
b g
B
2
73
g
B 84 g
H 123 g
g
Mo 17
N
197 s
s s
1
2
3