Genome organization, gene expression and germline development in C. elegans (with introduction to gene regulation)-Part A
advertisement

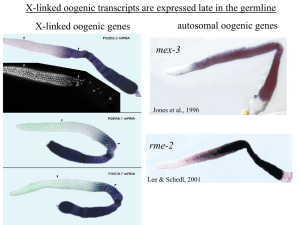
Genomes Organism Estimated size (in bases) Estimated gene # Average gene density/base Diploid chromosome # Human 2.9 x 109 ~30,000 1/100,000 46 Rat 2.8 x 109 ~30,000 1/100,000 42 Mouse 2.5 x 109 ~30,000 1/100,000 40 Drosophila 1.8 x 108 13,600 1/9,000 8 Arabidopsis 1.2 x 108 25,500 1/4,000 10 C. elegans 9.7 x 107 19,100 1/5,000 12 S. cerevisiae 1.2 x 107 6,300 1/2,000 32 E. coli 4.7 x 106 3,200 1/1400 1 H. influenzae 1.8 x 106 1,700 1/1000 1 http://www.ornl.gov Each cell within an organism contains a complete genome, but only deploys a fraction of the genes Positive: selective activation of a gene at a particular place and time to produce a gene product Negative: selective silencing of a gene or removal of a gene product at a particular place and time Gene expression regulation at the level of DNA Sequence-dependent cis-acting factors: promoters/regulatory sequences of genes trans-acting factors: proteins and RNAs that bind cis-elements and promote or repress gene expression DNA methylation: methylation of CpG islands promotes silencing Range: Usually functions at level of single gene, or at most a local group of genes Regulatory network from early sea urchin development Levine and Davidson, PNAS, 2005 DNA is embedded in chromatin Regulation of gene expression at the level of chromatin Sequence-independent linker histones: control DNA compaction and accessibility to trans-acting factors post-translational modifications of histone tails: control compaction of DNA and serve as docking sites for trans-acting factors Range: Can act at the level of a single gene, often acts over groups of genes and over larger domains (20-200kb), and can affect gene expression over an entire chromosome Regulation of gene expression at the level of RNA mRNA Stability/decay: length of poly A tail, binding of proteins and RNAs that either protect or degrade transcripts Subcellular localization: sequestration by proteins, ribosome stalling RNA also acts as a regulator of gene expression siRNA miRNA DNA is silenced at the level of histone modifications through an RNAi-like mechanism centromere repeats Genomics Technologies systematic gene mutation/RNAi screens sequence analyses Functional Networks genome-wide expression profiling genotyping global protein analysis nucleic acid/ protein interactions Genome organization is non-random with respect to gene expression in multiple organisms local chromatin chromosome Germ cells act to maintain the species somatic cells embryo somatic cells gametes embryo gametes C. elegans hermaphrodite germ line embryos sperm germline formation during larval development L1 L2 L3 L3/L4 late L4 stem cells meiosis young adult somatic gonad sperm oocytes Levels of gene regulation in the germline Chromosome: silencing of the X Large domain: clustering of germline-expressed genes Local domain: operon formation embryos sperm C. elegans DNA microarrays ~20,000 genes in the worm genome ~18,000 genes on the array Germline mutant comparisons wild type vs. no germ line (glp-4) sperm only (fem-3gf) vs. oocytes only (fem-1lf) C. elegans hermaphrodite germ line oogenic germline 3003 genes sperm 1380 genes Sperm genes are different from oogenic germline genes Large-scale in situ hybridization NextDB: Nematode EXpression paTtern DataBase Kohara lab Japan distal distal+ proximal 98% of all genes in oogenic germline category show germline expression by in situ proximal distal proximal Why? The oogenesis genes on the X chromosome express at lower levels than those on the autosomes A B The hermaphrodite paired X does not stain with antibodies against transcriptionally active chromatin conformation DNA Merge α -H3methylK4 diplotene stem cells pachytene diakinesis Hermaphrodite X is silenced early in meiotic prophase but not late diplotene A diakines inactive B Transgene 1 2 5 6 3 4 C D diakinesis -H3methylK4 diakines active tra







![Instructions for BLAST [alublast]](http://s3.studylib.net/store/data/007906582_2-a3f8cf4aeaa62a4a55316a3a3e74e798-300x300.png)