SUPPLEMENTARY MATERIAL Pathogenetics of Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins

SUPPLEMENTARY MATERIAL
Pathogenetics of Alveolar Capillary Dysplasia with Misalignment of Pulmonary
Veins
Przemyslaw Szafranski, Tomasz Gambin, Avinash V. Dharmadhikari, Kadir Caner
Akdemir, Shalini N. Jhangiani, Jennifer Schuette, Nihal Godiwala, Svetlana A. Yatsenko,
Jessica Sebastian, Suneeta Madan-Khetarpal, Urvashi Surti, Rosanna G. Abellar, David
A. Bateman, Ashley L. Wilson, Melinda H. Markham, Jill Slamon, Fernando Santos-
Simarro, María Palomares, Julián Nevado, Pablo Lapunzina, Brian Chung Hon-Yin,
Wong Wai-Lap, Yoyo Wing Yiu Chu, Gary Tsz Kin Mok, Eitan Kerem, Joel Reiter,
Namasivayam Ambalavanan, Scott A. Anderson, David R. Kelly, Joseph Shieh, Taryn C.
Rosenthal, Kristin Scheible, Laurie Steiner, M. Anwar Iqbal, Margaret L. McKinnon,
Sara Jane Hamilton, Kamilla Schlade-Bartusiak, Dawn English, Glenda Hendson,
Elizabeth R. Roeder, Thomas S. DeNapoli, Rebecca Okashah Littlejohn, Daynna J.
Wolff, Carol L. Wagner, Alison Yeung, David Francis, Elizabeth K. Fiorino, Morris
Edelman, Joyce Fox, Denise A. Hayes, Sandra Janssens, Elfride De Baere, Björn Menten,
Anne Loccufier, Lieve Vanwalleghem, Philippe Moerman, Yves Sznajer, Amy S. Lay,
Jennifer L. Kussmann, Jasneek Chawla, Diane J. Payton, Gael E. Phillips, Erwin Brosens,
Dick Tibboel, Annelies de Klein, Isabelle Maystadt, Richard Fisher, Neil Sebire, Alison
Male, Maya Chopra, Jason Pinner, Girvan Malcolm, Gregory Peters, Susan Arbuckle,
Melissa Lees, Zoe Mead, Oliver Quarrell, Richard Sayers, Martina Owens, Charles
Shaw-Smith, Janet Lioy, Eileen McKay, Nicole de Leeuw, Ilse Feenstra, Liesbeth Spruijt,
Frances Elmslie, Timothy Thiruchelvam, Carlos A. Bacino, Claire Langston, James R.
Lupski, Partha Sen, Edwina Popek, Paweł Stankiewicz
Author affiliations
P. Szafranski, T. Gambin, A. V. Dharmadhikari, S. N. Jhangiani, C. A. Bacino, J. R.
Lupski, P. Stankiewicz
Department of Molecular and Human Genetics, Baylor College of Medicine, Houston,
TX, USA
A. V. Dharmadhikari, P. Stankiewicz
Interdepartmental Program in Translational Biology and Molecular Medicine, Baylor
College of Medicine, Houston, TX, USA
P. Stankiewicz
Institute of Mother and Child, Warsaw, Poland
S. N. Jhangiani, J. R. Lupski
Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX, USA
J. R. Lupski
Department of Pediatrics, Baylor College of Medicine, Houston, TX, USA
K. C. Akdemir
Genomic Medicine Department, MD Anderson Cancer Center, Houston, TX, USA
J. Schuette
Division of Pediatric Anesthesia and Critical Care Medicine, Johns Hopkins Medical
Institutions, Baltimore, MD, USA
N. Godiwala
Division of Critical Care Medicine, Children’s National Health System, Washington, DC,
USA
S. A. Yatsenko, U. Surti
Department of Obstetrics, Gynecology, and Reproductive Sciences, Center for Medical
Genetics and Genomics, Magee-Womens Hospital of UPMC, Pittsburgh, PA, USA
Department of Pathology, University of Pittsburgh School of Medicine, Pittsburgh, PA,
USA
U. Surti
Department of Human Genetics, Graduate School of Public Health, University of
Pittsburgh, PA, USA
J. Sebastian, S. Madan-Khetarpal
Division of Medical Genetics, Children’s Hospital of Pittsburgh of UPMC, Pittsburgh,
PA, USA
R. G. Abellar
Department of Pathology, Columbia University Medical Center, New York, NY, USA
D. A. Bateman
Department of Pediatrics, Columbia University Medical Center, New York, NY, USA
A. L. Wilson
Children's Hospital of New York-Presbyterian, New York, NY, USA
M. H. Markham
Division of Neonatology, Division of Pediatrics, Vanderbilt University Medical Center,
Nashville, TN, USA
J. Slamon
Department of Obstetrics and Gynecology, Division of Maternal Fetal Medicine,
Vanderbilt University Medical Center, Nashville, TN, USA
F. Santos-Simarro M. Palomares, J. Nevado, P. Lapunzina
INGEMM, Instituto de Genética Médica y Molecular, IdiPAZ, Madrid, Spain
CIBERER, ISCIII, Madrid, Spain
B. H.-Y. Chung, W.-L. Wong, Y. W. Y. Chu, G. T. K. Mok
Department of Paediatrics and Adolescent Medicine, The University of Hong Kong,
Hong Kong, China
B. H.-Y. Chung
Department of Obstetrics and Gynaecology, and Centre for Genomic Sciences, The
University of Hong Kong, Hong Kong, China
E. Karem, J. Reiter
Pediatric Pulmonary Unit, Department of Pediatrics, Hadassah-Hebrew University
Medical Center, Jerusalem, Israel
N. Ambalavanan
Departments of Pediatrics, University of Alabama at Birmingham, Alabama, USA
Cell Developmental and Integrative Biology, University of Alabama at Birmingham,
Birmingham, AL, USA
S. A. Anderson
Department of Surgery, Division of Pediatric Surgery, University of Alabama at
Birmingham and Children's of Alabama, Birmingham, AL, USA
D. R. Kelly
Department of Pathology, University of Alabama at Birmingham and Pathology and
Laboratory Medicine Service, Children's of Alabama, Birmingham, AL, USA
J. Shieh
Division of Medical Genetics, Department of Pediatrics, and Institute for Human
Genetics, University of California San Francisco, San Francisco, CA, USA
T. C. Rosenthal
Genetics Department, Kaiser Permanente San Jose Medical Center, San Jose, CA, USA
K. Scheible
Department of Pediatrics, University of Rochester, Rochester, NY, USA
L. Steiner
Division of Neonatology, University of Rochester, Rochester, NY, USA
M. A. Iqbal
Pathology and Laboratory Medicine, University of Rochester Medical Center, Rochester,
NY, USA
M. L. McKinnon, S. J. Hamilton, K. Schlade-Bartusiak, D. English
Department of Medical Genetics, University of British Columbia, Vancouver, Canada
G. Hendson
Department of Pathology, University of British Columbia, Vancouver, Canada
E. R. Roeder, R. O. Littlejohn
Department of Pediatrics, Baylor College of Medicine, San Antonio, TX, USA
E. R. Roeder
Department of Molecular and Human Genetics, Baylor College of Medicine, San
Antonio, TX, USA
T. S. DeNapoli
Department of Pathology, Children’s Hospital of San Antonio, San Antonio, TX, USA
D. J. Wolff
Department of Pathology and Laboratory Medicine, Medical University of South
Carolina, Charleston, SC, USA
C. L. Wagner
Department of Pediatrics, Medical University of South Carolina, Charleston, SC, USA
A. Yeung, D. Francis
Victorian Clinical Genetics Services, Murdoch Childrens Research Institute, Parkville,
VIC, Australia
E. K. Fiorino
Division of Pediatric Pulmonary Medicine, The Children's Heart Center Steven and
Alexandra Cohen Children's Medical Center of New York, New York, NY, USA
M. Edelman
Division of Pediatric Pathology, The Children's Heart Center Steven and Alexandra
Cohen Children's Medical Center of New York, New York, NY, USA
J. Fox
Division of Medical Genetics, Steven and Alexandra Cohen Children's Medical Center of
New York, New Hyde Park, New York, NY, USA
D. A. Hayes
Pediatric Cardiology, The Children's Heart Center Steven and Alexandra Cohen
Children's Medical Center of New York, New York, NY, USA
S. Janssens, E. De Baere, B. Menten
Center for Medical Genetics, Ghent University and Ghent University Hospital, Ghent,
Belgium
A. Loccufier
Department of Obstetrics, Gynaecology, and Fertility, AZ St Jan Brugge, Brugge,
Begium
L. Vanwalleghem
Department of Anatomopathology, AZ St Jan Brugge, Brugge, Begium
P. Moerman
Department of Pathology UZ Leuven, Leuven, Belgium
Y. Sznajer
Center for Human Genetics, Cliniques Universitaires St-Luc, Universite Catholique de
Louvain, Brussels, Belgium
A. S. Lay
Division of Pediatric Cardiology, Children’s Mercy Hospital, Kansas City, MS, USA
J. L. Kussmann
Division of Clinical Genetics, Children’s Mercy Hospital, Kansas City, MS, USA
J. Chawla
Division of Paediatric Respiratory & Sleep Medicine, Lady Cilento Children’s Hospital,
Children’s Health Queensland Hospital and Health Service, Brisbane, QLD, Australia
The University of Queensland, Brisbane, QLD, Australia
D. J. Payton, G. E. Phillips
Division of Anatomical Pathology, Lady Cilento Children’s Hospital, Children’s Health
Queensland Hospital and Health Service, Brisbane, QLD, Australia
Pathology Queensland, Brisbane, QLD, Australia
E. Brosens, A. de Klein
Clinical Genetics Department, Erasmus MC-Sophia, Rotterdam, Netherlands
E. Brosens, D. Tibboel
Paediatric Surgery, Erasmus MC-Sophia, Rotterdam, Netherlands
I. Maystadt
Centre de Génétique Humaine, Institut de Pathologie et de Génétique, Gosselies, Belgium
R. Fisher
James Cook University Hospital, Middlesborough, UK
N. Sebire
Department of Paediatric Histopathology, Great Ormond Street Hospital for Children and
UCL Institute of Child Health, London, UK
A. Male, M. Lees
Clinical Genetics Unit, Great Ormond Street Hospital for Children and UCL Institute of
Child Health, London, UK
M. Chopra, J. Pinner,
Department of Medical Genomics, Royal Prince Alfred Hospital, Sydney, NSW,
Australia
G. Malcolm
Department of Newborn Care, Royal Prince Alfred Hospital, Sydney, NSW, Australia
G. Peters
Cytogenetics Department, The Children’s Hospital at Westmead, Westmead, NSW,
Australia
S. Arbuckle
Histopathology Department, The Children’s Hospital at Westmead, Westmead, NSW,
Australia
Z. Mead
Department of Histopathology, Addenbrooke’s NHS Trust Pathology Department,
Addenbrooke’s Hospital, Cambridge, UK
O. Quarrell, R. Sayers
Department of Clinical Genetics, Sheffield Children's Hospital, Sheffield, UK
M. Owens, C. Shaw-Smith
Molecular Genetics Department, Royal Devon and Exeter NHS Foundation Trust, Exeter,
UK
J. Lioy
Division of Neonatology, The Children's Hospital of Philadelphia, The University of
Pennsylvania, Perelman School of Medicine, Philadelphia, PA, USA
E. McKay
Department of Pathology, The Children's Hospital of Philadelphia, Philadelphia, PA,
USA
N. de Leeuw, I. Feenstra, L. Spruijt
Department of Human Genetics, Radboud University Medical Center, Nijmegen, the
Netherlands
F. Elmslie
South West Thames Regional Genetics Service, St George's University Hospital, London,
UK
T. Thiruchelvam
Critical Care and Cardiorespiratory Unit, Great Ormond Street Hospital NHS Trust,
London, UK
C. A. Bacino, J. R. Lupski
Texas Children’s Hospital, Houston, TX, USA
C. Langston, E. Popek
Department of Pathology and Immunology, Baylor College of Medicine, Houston, TX,
USA
P. Sen
Department of Pediatrics, Northwestern University, Chicago, IL, USA
METHODS
DNA isolation and sequencing
DNA was extracted from FFPE lung tissue or peripheral blood using MasterPure
Complete DNA & RNA Purification Kit (Epicentre, Madison, WI, USA) or Gentra
Purgene Kit (Qiagen, Germantown, MD, USA), respectively. PCR products were treated with ExoSAP-IT (USB, Cleveland, OH, USA), and directly sequenced by the Sanger method (Lone Star Labs, Houston, TX, USA). Sequences were assembled using
Sequencher v4.8 (GeneCodes, Ann Arbor, MI, USA).
RNA isolation and RT-qPCR
Gene expression was measured by estimating relative levels of mRNA. Total RNA was isolated from normal or transfected human foetal lung fibroblasts IMR-90 (ATCC,
Manassas, VA, USA) using miRNeasy Mini Kit (Qiagen, Hilden, Germany). RNA preps were treated with DNase using DNA-free Kit (Ambion, Austin, TX, USA), and converted to cDNA using SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA,
USA). RT-qPCR was performed using TaqMan Universal PCR Master Mix (Applied
Biosystems, Foster City, CA, USA) or Power SYBR Green kit (Applied Biosystems,
Warrington, UK). qPCR conditions included 40 cycles of 95°C for 15 sec and 60°C for 1 min.
For relative quantification of a transcript, the comparative CT method was used.
Cloning of the 16q24.1 region deleted in patient 122 and of the FOXF1 promoter
To determine whether the deleted region can function as a promoter of LINC01082 , we cloned the deleted 4.1 kb DNA fragment and measured its transcriptional activity in the
IMR-90 cell line. For comparison, we also cloned the 3 kb FOXF1 promoter region, which exhibits residual promoter activity in the absence of enhancers. The 4.1 kb noncoding fragment (chr16:86,216,561-86,220,676) deleted in patient 122.3, was amplified from the normal human DNA using the following LR-PCR primers: 5`-
AAGGTACCGGCATTTCTGTCACTCATTCAACAATCTGA-3`
AAGCTAGCGAGGTATGTTAGAGGAATAGAAGGACTGCCTTG-3`, and 5`which included the restriction sites for Kpn I and Nhe I, respectively. PCR was performed using
LA Taq DNA polymerase, applying 25 cycles of incubation at 94°C for 30 sec and at
68°C for 4 min. The amplified region was cut with
Kpn I and Nhe I, and cloned into Kpn I and Nhe I sites of the multiple cloning site (MCS) of the promoter-less vector pGL4.10
(Promega) to generate the pGL4.10.122 construct.
An ~ 5.5 kb 16q24.1 region containing the FOXF1 promoter and the FOXF1
ATG initiation codon (chr16:86,538,679–86,544,175) was amplified from the normal human DNA using the LR-PCR primers: 5`-
CTAGCTAGCACATTTCCTCATATTCTGTGTAGAGAGCACCT-3` and 5`-
TTGCGCCGATTCGAACGGGTGGCTGCTG-3` that included the restriction sites for
Nhe I and Bst BI, respectively. PCR was run using LA Taq DNA polymerase in the presence of 20% betaine, applying 25 cycles of incubation at 94°C for 30 sec and at 68°C for 5 min. The amplified FOXF1 promoter region was subsequently cut with Bst BI, blunt ends generated with DNA polymerase Klenow fragment, cut with Nhe I, and cloned into the Eco RV and Nhe I sites of the MCS of the pGL4.10 to generate the pGL4.10FOXpr construct.
Transcriptional activity assay
Transfections of the IMR-90 cells, were performed on the sub-confluent cells grown in
12-well plates. The IMR-90 cultures were maintained in the Eagle’s minimum essential medium (EMEM) supplemented with 2 mM L-glutamine and 10% FBS (ATCC). The cells were transfected with 1 μg of either pGL4.10.122, pGL4.10 (negative control), or pGL4.10FOXpr (positive control) using Lipofectamine 3000 with p3000 (Invitrogen).
Total RNA was prepared 48 h after transfection following lysis of the cells in Triazol, and converted to cDNA.
The transcriptional activity of the cloned fragments was assayed by qPCR by measuring relative quantity of Renilla luciferase ( Luc ) cDNA. At least three biological replicates for each construct were analysed. The Luc cDNA levels were normalized to cDNA levels of the pGL4.10 Amp gene. qPCR primers (lucF 5`-
GCACATATCGAGGTGGACATTA-3`, lucR 5`-CCACGATCCGATGGTTTGTAT-3`; ampF 5`-GCTGTCGTGATGCTAGAGTAAG-3`, ampR 5`-
AGAGTTGAACGAAGCCATACC-3`) were designed using PrimerQuest (IDT,
Coralville, IA, USA). cDNA synthesized using RNA isolated from IMR-90 cells transfected with the pGL4.10 plasmid was designated as a calibrator.
Bioinformatic analysis of the distant upstream enhancer region
Reference DNA sequences, chromatin modification, location of CpG islands, and ChIPseq data for the selected transcription regulators were accessed using the UCSC Genome
Browser (http://genome.ucsc.edu, GRCh37/hg19).
High-throughput chromosome conformation capture (Hi-C) (Lieberman-Aiden et al. 2009) interaction datasets for Lymphoblastoid cell line from B-lymphocytes
(GM12878), Human Umbilical Vein Endothelial Cells (HUVEC), Normal Human
Epidermal Keratinocytes (NHEK), and Human Mammary Epithelial Cells (HMEC), and human fetal lung fibroblasts (IMR-90) cell lines were downloaded from the GEO database (GSE63525). Normalized 25 kb resolution Hi-C interaction matrices of chromosome 16 for the aforementioned five cell lines were generated by multiplying
Knight and Ruiz normalization scores for two contacting loci and dividing raw observed values (MAPQGE30 filtered reads) at the interacting positions with this calculated normalization-score (Rao et al. 2014). IMR-90 cell line Pol2 ChIP-Seq was downloaded from UCSC Genome Browser ENCODE portal. Chromatin states calls (ChromHMM)
were downloaded from http://compbio.mit.edu/roadmap (REC 2015). HiCPlotter
(https://github.com/kcakdemir/HiCPlotter) was used to plot Hi-C data with chromatin states (Akdemir et al. 2015).
Microarray analyses
In addition to custom aCGH, a number of probands were studied using various microarrays: Illumina CytoSNP-850k BeadChip (Illumina, San Diego, CA, USA) (pt
120.3), Affymetrix Cytoscan 750K (pt 122.3), Affymetrix CytoScan HD array platform
(Affymetrix Inc., Santa Clara, CA, USA) (pts 125.3, 126.3, 135.3, and 136.3), 12x135K array (Roche-NimbleGen, Madison, WI, USA) (pt 127.3), ISCA v.1 180K (Agilent
Technologies, Santa Clara, CA, USA) (pts 128.3 and 133.3), and 4x180 K CGH-SNP array (Agilent Technologies) (pt. 139.3).
Whole exome sequencing
WES was performed at Baylor College of Medicine Human Genome Sequencing Center through the Baylor Hopkins Center for Mendelian Genomics as previously described (pts
114.3, 121.3) (Lupski et al. 2013) and at the University of California Los Angeles (pt
128.3) (Lee et al. 2014). WES in family 138 was performed as described [Reiter et al. manuscript submitted]. DNA sample, prepared into Illumina paired-end libraries, underwent whole exome capture using VCRome 2.1 design (Roche NimbleGen,
Madison, WI, USA), followed by sequencing on the Illumina HiSeq 2000 platform
(Illumina, San Diego, USA) with 78 paired-end reads coverage. Raw sequence data were post-processed using the Mercury pipeline (Reid et al. 2014), which performs conversion of raw sequencing data (bcl files) to the fastq format using Casava, mapping of the short reads against a human genome reference sequence (GRCh37) by the Burrows-Wheeler
Alignment (BWA), recalibration using GATK (McKenna et al. 2010), and variant calling using the Atlas2 suite (Challis et al. 2012). Variants were annotated using the in-housedeveloped “Cassandra” annotation pipeline (Bainbridge et al. 2011). Gene expression data were analysed through a gene annotation portal BioGPS (http://biogps.com).
To search for regions of absence of heterozygosity (AOH) in the WES data, we calculated B-allele frequency as a ratio of variants reads to total reads. These data were then processed using the Circular Binary Segmentation algorithm (CBS) (Olshen et al.
2004). To detect CNVs in families sequenced at BH-CMG (121, 124), we processed
WES data using CoNIFER software ( Krumm et al. 2012). Control set required by
CoNIFER was created based on WES data from 200 samples sequenced at BH-CMG using the same platform and processed with pipeline as ACDMPV families.
REFERENCES
Akdemir K, Chin L (2015) HiCPlotter integrates genomic data with interaction matrices.
Genome Biol 16:198
Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C,
Newsham I, Reid JG, Fink JK, Morgan MB, Gingras M-C, Muzny DM, Hoang
LD, Yousaf S, Lupski JR, Gibbs RA (2011) Whole-Genome Sequencing for
Optimized Patient Management. Sci Transl Med 3:87re3
Challis D, Yu J, Evani US, Jackson AR, Paithankar S, Coarfa C, Milosavljevic A, Gibbs
RA, Yu F (2012) An integrative variant analysis suite for whole exome nextgeneration sequencing data. BMC Bioinformatics 13:8
Krumm N, Sudmant PH, Ko A, O'Roak BJ, Malig M, Coe BP; NHLBI Exome
Sequencing Project, Quinlan AR, Nickerson DA, Eichler EE (2012) Copy number variation detection and genotyping from exome sequence data. Genome Res
22:1525-1532
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Das K, Toy T,
Harry B, Yourshaw M, Fox M, Fogel BL, Martinez-Agosto JA, Wong DA, Chang
VY, Shieh PB, Palmer CG, Dipple KM, Grody WW, Vilain E, Nelson SF (2014)
Clinical exome sequencing for genetic identification of rare Mendelian disorders.
JAMA 312:1880-1887
Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A,
Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender
MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES,
Dekker J (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289-293
Lupski JR, Gonzaga-Jauregui C, Yang Y, Bainbridge MN, Jhangiani S, Buhay CJ, Kovar
CL, Wang M, Hawes AC, Reid JG, Eng C, Muzny DM, Gibbs RA (2013) Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome
Med 5:57
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K,
Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis
Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297-1303
Olshen AB, Venkatraman ES, Lucito R, Wigler M (2004) Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5:557-572
Prothro SL, Plosa E, Markham M, Szafranski P, Stankiewicz P, Killen SA (2016)
Prenatal Diagnosis of Alveolar Capillary Dysplasia with Misalignment of
Pulmonary Veins.
J Pediatr 170:317-318
Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn
AL, Machol I, Omer AD, Lander ES, Aiden EL (2014) A 3D Map of the Human
Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell
159:1665-1680
Reid JG, Carroll A, Veeraraghavan N, Dahdouli M, Sundquist A, English A, Bainbridge
M, White S, Salerno W, Buhay C, Yu F, Muzny D, Daly R, Duyk G, Gibbs RA,
Boerwinkle E (2014) Launching genomics into the cloud: deployment of
Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics
15:30
Roadmap Epigenomics Consortium (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317-330
Supplemental Fig. S1 Differential methylation of a CpG island at the promoter region of the LINC01081 might contribute to epigenetic control of FOXF1 expression. The upper panel represents an example of array CGH plot of a deletion on maternal chromosome
16q24.1 (pt 119) that removed a portion of the FOXF1 upstream enhancer, including part of LINC01081 .
Supplemental Fig. S2 Comparison of transcriptional activities, of the basal FOXF1 promoter and the region deleted in patient 122.3 in fetal lung fibroblasts IMR-90.
Promoterless vector, pGL4.10, was used as a negative control.
Supplemental Fig. S3 Schematic representation of the proposed molecular mechanism of formation of the complex genomic rearrangement within the untranslated portion of the FOXF1 1 st
exon (pt 124.3). ( a ) The position of a copy the translocated region is indicated by a red bar. ( b ) The deleted fragment is shown as a red frame.
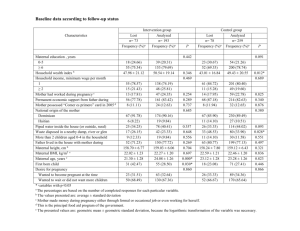
Supplemental Table S1 Genomic characteristics and parental origin of 42 reported and novel de novo deletion CNVs on chromosome 16q24.1 pathogenic for ACDMPV, and additional non-lung clinical features in ACDMPV patients. Thirteen novel cases are shaded
Patient
D1
Genomic coordinates
(GRh37/hg19)
Deletion size (kb)
85,148,264-
86,646,503
1,498
Location
Enhancer &
FOXF1
Breakpoints proximal distal
Alu Sq Alu Sq
Microhomology Mechanism of formation
17 bp MMBIR/FoSTeSx1
D2
D3
49.3 (D4)
D5
21.6 (D8)
-
-
-
-
90.3
104.3
85,717,653-
87,718,253
85,816,707-
86,719,506
85,845,070-
86,878,211
84,350,698-
87,847,575
86,600,342-
86,731,402
~85,890,261-
87,257,585
85,108,709-
86,720,212
~85,728,812-
86,831,579
~85,881,691-
87,882,985
81,094,654-
86,636,753
2,001
903
1,033
3,497
1,830
~1.370
1.612
~1.1
2.001
5,542
85,390,350/373-
87,964,597/620
2,574
Parental origin
Maternal
Cardiac
PDA
Clinical features in organs other than lungs
Gastrointestinal Genitourinary
Esophageal atresia, tracheoesophageal fistula, abnormal placement of anus
-
-
Dilated renal pelvices
Other
SUA
Reference
Stankiewicz et al. (2009)
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
L1ME1
Alu Sx
Alu Sx
Alu Sx
Alu Sx
ND
ND
ND
ND
FLAM_C
MER1
Alu Sx
Alu Sg1
Alu Sx
Alu Sx
ND
Enhancer &
FOXF1
Uniq AT-rich Alu Ya5
ND
ND
ND
Alu Sc5
4 bp MMBIR/FoSTeSx1 ND HLHS - Stankiewicz et al. (2009)
18 bp
8 bp
12 bp
43 bp
ND
ND
ND
ND
24 bp
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
ND
25 bp
AT-rich insertion
MMBIR/FoSTeSx1 or NHEJ
ND
ND
ND
MMBIR/FoSTeSx1
Maternal
Maternal
ND
Maternal
ND
ND
ND
ND
ND
Maternal rs2113236
ToF; PDA Duodenal atresia with annular pancreas, imperforate anus
-
Bilateral hydronephrosis, mod right, severe left
SUA
HLHS
IAA, dilated PA, large PDA, decreased size left ventricle
HLHS
-
Duodenal atresia with ileal cyst (Duodenal narrowing with massive dilation of the third and fourth portion of the duodenum; duplication of distal ileum and malrotation; stomach and initial part of intestines were malformed and tubular in nature). Annular pancreas
Right renal cyst extending into the abdomen.
Mild uretero-pelvic caliectasis
Bilateral renal pelviectasis
T11 butterfly vertebra, cleft lip, cleft palate, brachycephaly, SUA
Posterior rib fusions:
10/11 (right side),
9/10 and 11/12 (left side)
-
PDA ligated, structurally normal heart by echocardiogram
HLHS, hypoplasia of mitral valve and left ventricle, pulmonary valve atresia, subaortic VSD with overriding of aorta, PDA, proximal left PA stenosis,
PFO, and a persistent left superior vena cava
AVSD with a dysplastic tricuspid valve and two mitral valve leaflets. Small
PA
N/A
Adhesions between bowel loops, second part of duodenum and gallbladder, no malrotation seen at laparotomy
-
Annular pancreas with duodenal dilation proximal to the pancreas
N/A
-
Moderate to severe hydronephrosis, tortuous dilated ureters and thickened urinary bladder wall, suggesting possible posterior urethral valves.
Hypospadias
Bilateral dilation of the renal pelvocaliceal system with bilateral ureteral stenosis
Partial atrioventricular canal defect; ASD; VSD; PDA
ToF with severe right ventricular outflow tract obstruction
-
Pleural effusion, polyhydramnios. hypertelorism, broad flat nasal bridge. Hypotonia
-
N/A
-
Left kidney with moderate hydronephosis and hydroureter. Hypospadias.
Septated cystic hygroma, fetal hydrops, and SUA
(ultrasound). Limited autopsy
Prominent nasal bridge; deep-set eyes; mild retrognathia. segmental abnormality of T10 vertebral body
-
-
Prenatal. SUA. Trunk and head edema. Thickened nuchal fold (3.7 mm).
Stankiewicz et al. (2009)
Stankiewicz et al. (2009)
Stankiewicz et al. (2009)
Stankiewicz et al. (2009)
Yu et al. (2010)
Zufferey et al (2011)
Garabedian et al. (2012)
Handrigan et al. (2013)
Sen et al. (2013b)
Sen et al. (2013b)
118.3 86.027,015/033-
87,198,727/745
1,172 Enhancer &
FOXF1
Alu Sx Alu Jb
120.3
125.3
133.3
86,010,039/061-
87,847,731/753
1,838
86,182,703/738-
87,651,485/531
1,469
83,666,375-
87,300,195
3,634
Enhancer &
FOXF1
Enhancer &
FOXF1
Enhancer &
FOXF1
Alu
Alu
Y
Y
MER41B
Alu
Alu Jb
Alu
Y
Y
135.3
140.3
D6
~85,728,498-
87,322,987
~86,220,144-
87,277,604
86,493,531-
86,596,413
~1,596
~1,057
103
Enhancer &
FOXF1
Enhancer &
FOXF1
FOXF1
ND
ND
Alu Y
115.3 86,505,450-
86,575,461
70 FOXF1
ND
ND
Alu Y unique unique
129.3
122.3
86,545,491-
86,546,265
86,216,561-
86,220,676
1
4
Intronic
Enhancer
28.7 (D10) ~86,140,499-
86,285,499
~145 Enhancer
Unique unique
Unique unique
ND ND
19 bp
21 bp
35 bp
Absent
ND
ND
19 bp
41.4
59.4
94.3
~86,530,499-
86,541,499
86,539,362-
86,554,368
86,541,294-
86,690,004
~11
15
149
FENDRR , promoter
FOXF1
FOXF1
ND ND
Low complexity
GC-rich
Unique
SINE MIR hAT-
Charlie1a
ND
Absent
Absent
Absent
Absent
Absent
ND
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
NHEJ or
MMBIR/FoSTeSx1
ND
ND
MMBIR/FoSTeSx1
ND
ND AVSD Suspected bowel obstruction (large loop persistent prenatally). Not evaluated postnatally.
Intestinal malrotation Maternal 15xGA microsatellite,
86,256,052/82
Maternal 15xGA microsatellite,
86,231,351/80
Maternal rs1704502
Aortic coarctation with arch hypoplasia and a bicuspid aortic valve
AVSD
Coarctation of aorta.
Aneurysmal dilatation of
PDA, PFO, Small mitral valve, bicuspid aortic valve, persistent left superior vena cava with suggestion of noncompaction cardiomyopathy
Coarctation of aorta Maternal
SNP array
Maternal,
SNP array
ND
-
-
Malrotation with in utero perforation, annular pancreas with duodenal stenosis/constriction
-
-
AVSD, ostium primum ASD, small VSD, PDA, mild
HLHS, bicuspid AV
Possible duodenal atresia, possible intestinal malrotation
ND N/A N/A
-
-
Bilateral hydroureter and bilateral hydronephrosis
-
Hydronephrosis
Bilateral hydronephrosis
Prenatal
Bilateral cerebellar heterotopia
-
Hypoglycaemia, SUA, deep set eyes with posterior nuchal webbing, bilateral choroid plexus cysts
Prenatal. Hygroma colli.
SUA
Prenatal
Moderate bilateral pelvicaliectasis, mild to moderate proximal urethral dilatation, probable mild to moderate distal ureteral dilatation
N/A
Absent spleen, transverse orientation of the liver, compatible with abdominal heterotaxy
N/A
Prothro et al. (2015) this study this study this study this study this study
Stankiewicz et al. (2009)
Szafranski et al. (2013a)
NHEJ or
MMBIR/FoSTeSx1
Maternal Large ASD, PDA - - - Szafranski et al. (2013a)
NHEJ or
MMBIR/FoSTeSx1
NHEJ or
MMBIR/FoSTeSx1
Maternal
Maternal
NHEJ or
MMBIR/FoSTeSx1
NHEJ or
MMBIR/FoSTeSx1
ND
Maternal
Paternal rs8061995 rs12444199 rs276991
Maternal
Hypoplastic aortic arch.
Large PDA (which allowed perfusion to the descending aorta and left subclaviar artery; ASD and VSD.
-
Intestinal malrotation
Imperforate anus (low rectal atresia), marked malrotation of the large intestines with loss of peritoneal fixation, shortened mesenteric root with kinking and luminal narrowing of the small intestine.
N/A N/A
Bilateral hydronephrosis
-
N/A
PDA
Small PDA
-
-
-
- none
-
N/A
-
-
Sen et al. (2013b) this study
Szafranski et al. (2013b) this study
Stankiewicz et al. (2009)
47.4 (D9) 85,867,768-
86,392,161
524
57.3
60.4
64.5
77.3
81.3
82,014,639/716-
86,300,403/481
83,673,382/476-
86,298,284/378
86,147,527/566-
86,287,120/159
86,212,041/067-
86,448,132/158
~86,194,972/195
,808-
86,354,712/355,
161
4,286
2,625
140
236
~160
Enhancer
Enhancer
Enhancer
Enhancer
Enhancer
Enhancer
ND ND
LINE L1
LINE L1
Alu Sz
Alu Sc
ND
LINE L1
LINE L1
Alu Sx
Alu Sq2
ND
95.3
96.3
86,118,131/141-
86,287,054/064
85,979,487-
86,361,223
169
382
Enhancer
Enhancer
Alu Jb
Unique
Alu Sx
Unique
99.3
111.3
117.3
119.3
126.3
84,764,628/647-
86,238,601/620
86,077,955/958-
86,271,915/918
86,055,159/200-
86,288,226/267
1,474
194
233
86,148, 250-
86,301,591
Insertion at the
BP: GCACGCA
84,875,483/490-
86,386,861/868
153
1,511
Enhancer
Enhancer
Enhancer
Enhancer
Enhancer
Alu Sc8
LTR ERVL
LTR16C
Alu Sp
Alu Y
LINE
L1PA3
Alu Sp
Alu
Alu
Y
Sx1
LINE
L1PA2
Alu Sx
127.3 86,209,157/194-
86,301,558/595
92 Enhancer
136.3
139.3
~85,146,556-
86,393,283
~85,877,026-
86,282,104
~1,250
~405
Enhancer
Enhancer
ND, not determined; BP, breakpoint
L1PA5
ND
ND
L1PA2
ND
ND
1 bp
77 bp
96 bp
41 bp
28 bp
ND
12 bp
Absent
19 bp
3 bp
41 bp
Absent
7 bp
37 bp
ND
ND
MMBIR/FoSTeSx1 or NHEJ
NAHR or
MMBIR/FoSTeSx1
NAHR or
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
ND
MMBIR/FoSTeSx1
NHEJ or
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
NAHR or
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
MMBIR/FoSTeSx1
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal
Maternal 15xGA microsatellite,
86,231,351/80
Maternal 15xGA microsatellite,
86,256,052/82
-
-
N/A
N/A
-
PDA with a right to left shunt, small pericardial effusion, right ventricular dilatation and hypertrophy, mitral valve had a parachute configuration with mild dilation of the left ventricle
Significant R/L shunt. Open ductus Botalli. Normal pulmonary vein connections
Small muscular VSD
Suspected intestinal malrotation, imperforate anus
-
N/A
N/A
-
-
-
-
PDA, dilated right ventricle with depressed function
-
Omphalocele with a diameter of 6 cm only intestines present, no involvement of liver.
-
-
-
Bicornuate uterus with cervical duplication
-
-
-
-
N/A
N/A
-
-
-
Bilateral hydronephrosis due to urethra valves
Multiple butterfly vertebrae Stankiewicz et al. (2009)
-
N/A
N/A
-
Unilobar left lung
-
-
-
-
Limited autopsy
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2013a)
Szafranski et al. (2014)
Szafranski et al. (2014) this study
- - - - this study
MMBIR/FoSTeSx1 Maternal 15xGA microsatellite,
86,231,351/80
NAHR or
MMBIR/FoSTeSx1
ND
ND
Maternal 15xGA microsatellite,
86,256,052/82
Maternal
SNP array
Maternal rs190442964
Balanced AVSD
PDA and PFO
-
-
Intestinal malrotation
-
-
-
Uterus didelphys
Mildly dilated right ureter
-
-
-
-
-
-
- this study this study this study this study