26. Replication.doc
advertisement

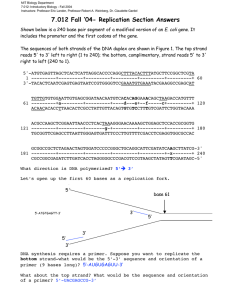
D’YOUVILLE COLLEGE BIOLOGY 102 - INTRODUCTORY BIOLOGY II LECTURE # 26 DNA REPLICATION 1. Basic Outline: (ppt. 1) REPLICATION DNA -----------------------> 2 identical DNAs (in nucleus) TRANSCRIPTION DNA -----------------------> mRNA (& other RNAs) (in nucleus) TRANSLATION mRNA -----------------------> protein (at ribosome) • base sequence of DNA codes for amino acid sequence of protein; one gene codes for one protein 2. DNA as Hereditary Material: • genes on chromosomes (Morgan's X-linkage studies); chromosomes made of nucleoprotein; thus genes must be nucleic acid or protein • “Transforming principle”: Griffith experiment with S & R strains of Pneumococcus (fig. 16 – 2 & ppt. 2); infection with S: lethal, mice died; infection with R: harmless, mice lived; infection with killed S: harmless, mice lived; infection with R + killed S: lethal; something from killed S rendered R infective (live S cells were found in blood of dead mouse) • Avery, MacLeod, & McCarty identified transforming principle as DNA; killed S cells were treated in separate experiments to distinguish between DNA, RNA or protein as transforming principle; only when DNA was not inactivated did transformation occur • DNA was confirmed as genetic material in virus infection experiment (Hershey & Chase); 35S-labeled protein & 32P-labeled DNA study (fig. 16 – 4 & ppt. 3) Bio 102, spr. 2013 3. lec. 25 - p. 2 Double Helix & Template Theory: • DNA structure elucidated by Watson & Crick (fig. 16 – 1 & ppt. 4) (modeled X-ray diffraction data of Wilkins & Franklin) (fig. 16 – 6 & ppt. 5) • Chargaff showed species-specific differences in DNA base content; A = T, C = G (Chargaff’s rules) • two antiparallel polynucleotide strands arranged in double helix, Hbonded by complementary base pairing (A - T; C - G) (figs 16 – 5, 16 – 7, 16 – 8 & ppts. 6 - 9); this model explained Chargaff’s rules • each strand = template for its complementary strand 4. Replication of DNA (template copy mechanism) (fig. 16 - 9 & ppt. 10): • semiconservative replication (figs. 16 –10 & ppt. 11); 14N15N DNA study (Meselson & Stahl) (fig. 16 – 11 & ppt. 12) • mechanisms: replication origins - base sequences recognized by enzymes which open double strand replication bubbles (replication forks at each end) (fig. 16 – 12 & ppt. 13) - helix opened up by combined action of helicase & single strand binding proteins (fig. 16 – 13) Bio 102, spr. 2013 lec. 25 - p. 3 - DNA polymerases add nucleotides in 5’ to 3’ direction (fig. 16 – 14 & ppt. 14) - antiparallel strands result in continuous synthesis (leading strand) on one template & discontinuous synthesis (lagging strand) on the complementary template (Okazaki fragments so formed are joined by DNA ligase) (figs. 16 – 15, 16 – 16 & ppt. 15) - primer RNA laid down by primase (figs 16 – 15 to 16 – 17 & ppt. 16) - summary of replication (fig. 16 - 17, table 16 - 1 & ppts. 17 & 18) - errors corrected by repair enzymes (fig. 16 – 18 & ppt. 19)