Genomic DNA was isolated from venous blood using the ... Frankfurt, Germany) according to the manufacturer’s instructions. Array-CGH was carried... SUPPLEMENTARY DATA
advertisement

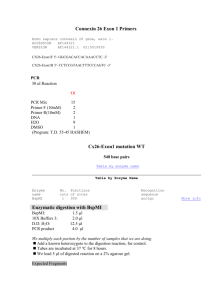
SUPPLEMENTARY DATA MATERIALS AND METHODS Array-based comparative genomic hybridization (CGH) Genomic DNA was isolated from venous blood using the QIAamp DNA Mini Kit (Qiagen, Frankfurt, Germany) according to the manufacturer’s instructions. Array-CGH was carried out using the 1x244K platform from Agilent (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer’s instructions. Homozygosity mapping Genomic DNA was isolated from venous blood using the Puregene® DNA purification kit (Gentra Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions. Genotyping was performed using the Mapping 250K Nsp array from Affymetrix containing 262264 SNPs (analysis performed by DNA Vision, Gosselies, Belgium). Genomic regions harbouring 25 consecutive homozygous SNPs (approximately 250 kb) or more were marked. To overcome SNP miscalls, regions spanning maximum 50 kb between two marked regions were considered homozygous. Eventually, regions that contained at least 100 consecutive homozygous SNPs were assigned as a candidate region if they were homozygous for the same allele in all affected patients and had a different genotype in all non-affected individuals. Mutation screening PCR amplification was performed with 50ng genomic DNA, 0.6 µM of each primer (oligonucleotide primer pairs (Invitrogen, Carlsbad, CA, USA) are listed in Supplementary Table 1), 1 mM MgCl2, 0.12 mM dNTP mix (Invitrogen), and 0.5 units of KAPATaq HotStart DNA polymerase in 1 x KAPATaq HotStart Buffer (Kapa Biosystems, Woburn, MA, USA). Thermal cycling conditions consisted of an initial denaturation step of 94°C for 4 minutes, 12 cycles of 94°C for 20s, 62°C for 15s, and 72°C for 60s with a decrease in annealing temperature of 1°C each cycle, followed by 24 cycles of 94°C for 40s, 50°C for 40s, and 72°C for 30s, and a final extension at 72°C for 10 minutes. PCR products were bidirectionally sequenced using the 1 BigDye® Terminator Cycle Sequencing kit protocol (Applied Biosystems, Foster City, CA, USA), followed by analysis on the ABI 3730XL Genetic Analyzer (Applied Biosystems). The presence of the mutation was confirmed by digesting the amplification products, generated with primers RIN2_Exon8_1_F and Exon8_2_R (Supplementary Table 1), with the restriction endonuclease AlwNI (10 units each reaction, New England Biolabs, Ipswich, MA, USA) for 2 h at 37°C. The digested fragments were separated on a 2% agarose gel (Invitrogen), and visualised with ethidium bromide (Sigma-Aldrich, St. Louis, MO, USA). Quantitative real-time PCR Fibroblasts of patient PIII.2 and three non-related control individuals were cultured during 16-18 hours in the absence or presence of 5 mg cycloheximide (Sigma-Aldrich) dissolved in 100 µl 100% ethanol (ThermoFisher Scientific, Waltham, MA, USA) (Carter et al. 1995). Total RNA was extracted using the RNeasy® mini kit (Qiagen) according to the manufacturer’s instructions, followed by DNase treatment using RQ1 RNase-free DNase (1U/µl) (Promega, Madison, WI, USA) and a desalting step using Microcon-100 spin columns (Millipore, Billerica, MA, USA). cDNA was synthesized from 2 µg of DNase treated RNA with the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s recommendations. qPCR amplification mixtures contained 20 ng template cDNA, 1x SYBR Green I Master Mix buffer (Applied Biosystems) and 250 nM forward (5’-GGGTCTTGTATCCACGTAAC-3’) and reverse (5’-AGGTACAAAGCCAACCAAAT-3’) primer (Biolegio, Nijmegen, The Netherlands). Reactions were run on the LightCycler 480 (Roche, Pleasanton, CA, USA). The cycling conditions consisted of a 5-minute pre-incubation at 95°C and 45 cycles at 95°C for 10 seconds, 60°C for 30 sec and 72°C for 1 sec. Normalization was performed using the reference genes HPRT1 (RT Primer DB ID: 5; http://medgen.ugent.be/rtprimerdb/) and YWHAZ (RT Primer DB ID: 9). Analysis of the data was performed with qBasePlus software (Biogazelle, Ghent, Belgium). For the patient and each individual, total RNA was isolated from two independent fibroblast cultures and each of these samples was analyzed in triplicate during a single qPCR assay. In order to investigate the presence of a shortened alternative transcript that lacks the 306 bp mutant exon, PCR amplification was performed as described above using three oligonucleotide 2 primer pairs: pair 1 forward 5’-ATGACCCAGGTCAAGAACTA-3’ and reverse 5’CAAGAAGTCATCAGCGCCATA-3’, pair 2 forward 5’-ATGACCCAGGTCAAGAACTA-3’ and reverse 5’-GCTGCTTGTTCTTCTTGGAAAT-3’ and pair 3 forward 5’- GGTGCTTAGTGCAGGACTAC-3’ and reverse 5’-GCTGCTTGTTCTTCTTGGAAAT-3’. Thermal cycling conditions consisted of an initial denaturation step of 95°C for 2 minutes, 14 cycles of 95°C for 30s, 60.5°C for 30s for pair 1 (59.7°C for pair 2 and 61.8°C for pair 3), and 72°C for 70s with a decrease in annealing temperature of 0.5°C each cycle, followed by 19 cycles of 95°C for 30s, 53.5°C for 30s for pair 1 (52.7°C for pair 2 and 54.8°C for pair 3), and 72°C for 70s, and a final extension at 72°C for 5 minutes. Amplified fragments were analyzed on the LabChip®GX (Caliper Life Sciences, Hopkinton, MA, USA). 3 Supplementary Table 1 RIN2 sequencing oligonucleotide primer pairs Amplicon Forward primer Reverse primer Exon 1 gtgagtgttctgtggcttag tcagagagcagacttccct Exon 2 ctattgaaaagcctgagttccc tgcaagtcagcccttgg Exon 3 gggacggtctccttgttag tttaagccggcagcagt Exon 4 gatgcttgggttccaaca gctccttccatttccactc Exon 5 tcctgtcccagaacgtg atccttcaagaaacctcaggtta Exon 6 gtatggtgtctacttaggacattt tcagtgagcccaccattg Exon 7_1 tgaaggaatggtctgagtgt ttgactgtttctggcatgc Exon 7_2 ggctggccaggactgaa ccatggtctcttggtcact Exon 7_3 cagcagcctggaggact gttgcactgcccggaca Exon 8_1 cttggtgttgtctgttggt attttctccacatccacaaaatca Exon 8_2 gaagcaactcaaggagaacc cagagaatcacttgtagttctgtaa Exon 9 agcaagaaggatgtgaaagaaag aatgttatgtgacatgaatttcgt Exon 10 gctggcatcccctgtta cctctgagctgggatgt Exon 11 cccatcatgctcactatctagtt tttggatgcaccactggg 4 Supplementary Fig. 1 Illustration of the evolution of the facial dysmorphism of patient PIII.2 at age 14 (a and b) and age18 years (c and d). (Informed consent was obtained from the patients for publication of this report and these photographs) 5 Supplementary Fig. 2 Illustration of the evolution of the facial dysmorphism of patient PIII.4 at age 5 (a and b), age 8 (c and d) and age 12 (e and f). (Informed consent was obtained from the patients for publication of this report and these photographs) 6 Reference Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, Wilkinson MF (1995) A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem 270: 28995-9003 7