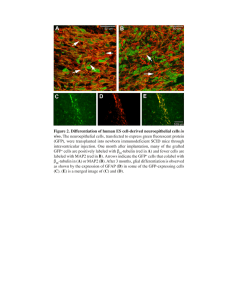
Development of the Moss Assessment in Space Physcomitrella patens Virginia Slater
advertisement

Development of the Moss Physcomitrella patens for Assessment in Space Virginia Slater Kirk Findlay, TJ White, Dr. Maria Ivanchenko and Dr. Terri Lomax Role of Plants on Earth The Role of Plants in Space • • • • • Life Support Food Medicines Water Recycling Waste Recycling Exploration…… …..colonization….. …..and habitation The Unique Conditions of a Space Environment • • • • • Microgravity Cosmic Radiation Low Atmospheric Pressure High CO2 Temperature Microarray Technology • Measures temporal and spatial gene expression. • Provides information of gene involvement in a process or pathway. Microarray Technology Microarray Technology Microarray Technology Microarray Technology NASA Free-Flyer Plant Genetic Assessment and Control Module Resource Inputs: Resource Adjustments: Biological Plants Seed/spores Microbes Biological Activate Genes Deactivate Genes Alter Genes Physical Light Water Gases Nutrients Plant-Centered Life-Support System Life Support Outputs: Food Medicines Vitamins Gas Recycling Water Recycling Waste Recycling Aesthetics/Avocation Physical Change light Change water Change gases Change nutrients Objective • Design plants to be used to assess the NASA Fundamental Space Biology FreeFlyer Satellite Program. • Design and produce genetic constructs that are coupled with fluorescent tags. • Transform the constructs into the moss Physcomitrella patens. • Monitor protein expression to establish baseline standards. Why Moss? • Physcomatrella patens is a model organism for the study of plant development. Why Moss? • Protonema – Cell division and growth. – Signal transduction • Gametophore – Organogenesis – Establishment of the body plan. – Plant development Why Moss? • Physcomatrella patens is a model organism for the study of plant development. • Contains a super efficient gene targeting system. – 90% efficiency Hypothesis • Rubisco is located in the chloroplasts. • We hypothesize that the Rubisco/GFP will cause the chloroplasts to fluoresce producing a signal that will be detectable from ground control. Methodology • Design primers for a portion of the Rubisco gene. • Use designed primers in PCR to amplify that portion of Rubisco. • Restrict the moss PCR product (Rubisco). • Restrict plasmid pMBL5. • Ligate pMBL5 and the Rubisco fragment together. • Repeat with GFP (next slide). Methodology Not I isco Plasmid DNA Bam HI Kpn I GFP Homologous Recombination Genomic DNA Rub isco isco Plasmid DNA GFP Homologous Recombination Stop Codon Rub isco Recombinant DNA GFP isco Results 1 kb Ladder pMBL5 and GFP fragment pMBL5 w/o insert pMBL5 and Rubisco fragment 1st Cut Results 1 kb Ladder pMBL5 and GFP fragment pMBL5 w/o insert pMBL5 and Rubisco fragment 2nd Cut Additional Strategy • Repeat the experiment using a structural protein. Digestion of Expansin 1 kb Ladder pMBL5 BamH I/Kpn GFP fragment - BamH I/Kpn I ?!?!?!? pMBL5 Not I/BamH I Expansin fragment - Not I/BamH I Expansin fragment - Not I/BamH I PP Expansin Gene AGATCTAACCACGAGAGTTTGGTGTGATCTCTGCAGTTTAAGCTAGTGAGCTGGTAGCAAGGGGCGATGGCGAG GCATAATGCAACAAAGCCTGTGACACTCATTCTTGCTGCACTGATGGTTCTTTCAGCCACCGACAACGTCGAAG GTCGTCATCACGTCAGAGATGGAAAAAACTGGCGCAAAGCTCATGCAACTTTCTACGGGGGTGCTGATGCTTCA GGAACTATGGGTAACTTTTTTCAACCTCTTGTTCAACTTCGGAGGCGTCCCATGAATCCTTACAAGTAGTAATT AAAACTTAAGTTTCTTGACAATGTGTATGCTTCCTATCTATTTGGAACTAAACCATTCCTCTGCTCTGATCAGA AACTTGAATCAGCTGCAAAGAGAATAAGACGCCAATATACATGTATCAGAAAACTAACGAAGAGGACTACAAAT TTTGGATCTCTTCCATGTAGCTCTTGTCCATAAGGCACCACCTTATGGAGAAATTTTTTTTCGAAAGTTTTGAA TTCAGAGATCGGTTGACTAAATAGTAACCTTCGAATGTGCAGACGGTGCATGCGGTGCATGCGGTTACGGAAAC CTCTACAGCACTGGCTATGGAGTCGATTCGACAGCTTTGAGTACAGCTCTTTTCAACAATGGGGCAAAATGCGG AGCTTGTTTTGCGATCCAATGCTATCGTTCACAGTATTGCGTTCCAGGTTCACCTGTAATCACTGTCACAGCTA CAAACTTCTGCCCTCCAACCACAAAGGTGATGGCACGCCAGGATGGTGTAATCCGCCAATGCGTCACTTCGACC TTGCGCAGCCTAGCTTCACCAAAATCGCTAAGTATAGAGCCGGCATCGTCCCCGTTCTCTTCAGAAGGTGTGCA TTGCGTTGAAGACTGATTTGTAAATTGTGACTTTAAGCCTTAATTACTGAGGATGGAGACAGCTGTGCAATCAC TTCGCAAATTAAACCATGCATGTTTTTAAGAAAACAGAAACGGCAGAACAAACCGTCAGCCAATTGAAAGAGAT GTTCTGAAACTTAGTAAAAGAGGTTGTCTTAGTCCTGTTTGGATTGGTAGTTTGATATTACGAAGTCCGTACTG ACCAAAACTTTGTTATATGCCTTGCACAATGCAGGGTACCATGCGAGAAAAAAGGTGGCGTCAGGTTCACTATC AATGGAAATAAGTATTTCAATCTCGTCCTAGTTCACAATGTTGGTGGAAAAGGCGATGTGCACGCAGTAGACAT AATACAGAATGGATTCCCATGAAGCGAAACTGGGGAATGAACTGGCAACAGATGCT GTTATGACCAAG GGATCC TGGCCAGGCACTCTCCTTCCGAGTGACAACCAGTGATGGTAAGACCATAGTCTCTATGAACGCAACGCCATCTC ACTGGAGCTTCGGCCAGACCTTCGAGGGAGGTCAGTTCGCTATGAATTGAATTCTGTAACCCCAAGAGCGGTGC CACTCGATGAATGCTTTAGGCGAAGAGTTGATCCACAAGGGAACCTAGACGCAGTTGAGTCTCAATCTAGCTTC ATGATATTTGTTGATACCTATACTGGATACCAATCGGCGTCTTGAATCCTCAACATCTACCCTCCACGCTTTAC CCAGATATCCCGAGATCTGGCCAACGTGAACGGTTTTGAAATTTACCAATATCAGTAGCACATAAAACCCATGG GGTACAATGATTTGTAGGTAGTGCGCAATCATGGGGAT Future Plans • Transform the moss cultures and select for stable transformations. • Repeat the experiment using a structural protein. • Monitor and analyze proteins levels to establish baseline standards for ground plants. • Test moss cultures in space. Acknowledgements • Howard Hughes Medical Insitute • McNair Scholar’s Program • Dr. Terri Lomax • Dr. Maria Ivanchenko • Kirk Findlay • TJ White • Dr. Kevin Ahern • Dr. Indira Rajagopal • The Lomax Lab • HHMI and McNair Scholars • Dept. of Botany and Plant Pathology