Multiple Linear Regression
advertisement

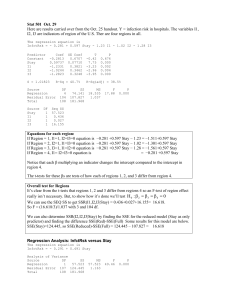
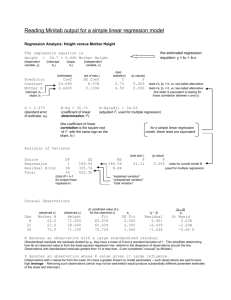
Multiple Linear Regression Extend model to many explanatory variables • Fitting as before, output essentially the same • F-test for overall significance • Individual t-tests for each variable – depends on the order of inclusion in the model – Explanatory variables can be highly related 1 Multiple Regression - Example Regression Analysis: edible versus height, width, length, shell The regression equation is edible = - 10.2 + 0.144 height + 0.204 width - 0.0199 length + 0.0884 shell Predictor Constant height width length shell Coef -10.189 0.14350 0.2043 -0.01989 0.08843 S = 4.04328 SE Coef 4.640 0.07481 0.1600 0.03712 0.01550 R-Sq = 88.7% T -2.20 1.92 1.28 -0.54 5.71 P 0.031 0.059 0.206 0.594 0.000 R-Sq(adj) = 88.1% 2 ANOVA – sums of squares Analysis of Variance Source Regression Residual Error Total Source height width length shell DF 1 1 1 1 DF 4 77 81 SS 9874.0 1258.8 11132.8 Seq SS 8631.4 710.3 0.1 532.1 MS 2468.5 16.3 F 151.00 Source shell length width height P 0.000 DF 1 1 1 1 Order of terms matters! Seq SS 9671.1 95.1 47.6 60.2 3 Variable Selection Methods • Best subsets – looks at all possible selections of explanatory variables • Stepwise – Forward – add in variables one at time – Backward – start with full model and remove insignificant variables – Full stepwise – combination of forward and backward 4 Best subsets Response is edible Vars 1 2 3 4 R-Sq 86.9 88.5 88.7 88.7 R-Sq(adj) 86.7 88.2 88.2 88.1 Mallows C-p 11.4 2.6 3.3 5.0 S 4.2744 4.0339 4.0248 4.0433 h e i g h t w i d t h l e n g t h s h e l l X X X X X X X X X X Look for maximum R-sq(adj) and minimum C-p 5 The regression equation is edible = - 9.81 + 0.0994 shell + 0.161 height Predictor Constant shell height Coef -9.813 0.09939 0.16070 S = 4.03386 SE Coef 4.402 0.01150 0.04884 R-Sq = 88.5% T -2.23 8.64 3.29 P 0.029 0.000 0.001 R-Sq(adj) = 88.2% Analysis of Variance Source Regression Residual Error Total DF 2 79 81 SS 9847.3 1285.5 11132.8 MS 4923.6 16.3 F 302.58 P 0.000 6 Forward Selection Response is edible on 4 predictors, with N = 82 Step 1 2 Constant 4.423 -9.813 shell T-Value P-Value 0.1327 23.01 0.000 height T-Value P-Value S R-Sq R-Sq(adj) Mallows C-p 0.0994 8.64 0.000 0.161 3.29 0.001 4.27 86.87 86.71 11.4 Leads to same model Shell + height 4.03 88.45 88.16 2.6 7 Model Checking – not great! Residual Plots for edible Normal Probability Plot of the Residuals Residuals Versus the Fitted Values 99 10 90 5 Residual Percent 99.9 50 10 -5 -10 1 0.1 0 -10 -5 0 Residual 5 10 0 Histogram of the Residuals 12 24 36 Fitted Value 48 Residuals Versus the Order of the Data 40 Residual Frequency 10 30 20 10 5 0 -5 -10 0 -12 -6 0 Residual 6 12 1 10 20 30 40 50 60 Observation Order 70 80 8 Categorical Data; Factors • Categorical variables defined as factors are not handled automatically by regression routines => need to transform to dummy variables ! • A k-category factor has – k levels – unique labels for each level – Use Make Indicator Variables to create dummy 0/1 indicators 9 Factor - example • Create 0/1 indicators for SpeciesGroup – Name them oyster, mussel • Tally for Discrete Variables: SpeciesGroup, oyster, mussel SpeciesGroup M O N= Count 92 81 173 mussel Count 0 81 1 92 N= 173 oyster 0 1 N= Count 92 81 173 10 Comparing Two Groups • Simply fit a regression model with a dummy variable for the two-level-factor as covariate. • Regression analysis is equivalent to doing a 2-sample t-test with equal variances 11 Regression Output The regression equation is Cadmium = 0,166 + 0,219 oyster Predictor Constant oyster • • • Coef 0.16562 0.21919 SE Coef 0.01286 0.01876 T 12.87 11.68 P 0.000 0.000 The intercept is the mean for the first level of the factor (M) The oyster estimate is the difference between the means The t-value tests whether this difference is significantly different from zero, i.e. if the two means are equal 12 t-test output Two-sample T for Cadmium SpeciesGroup N Mean StDev M 89 0.1656 0.0870 O 79 0.385 0.151 SE Mean 0.0092 0.017 Difference = mu (M) - mu (O) Estimate for difference: -0.219192 95% CI for difference: (-0.256232, -0.182152) T-Test of difference = 0 (vs not =): T-Value = -11.68 P-Value = 0.000 DF = 166 Both use Pooled StDev = 0.1214 13 ANOVA table: Cadmium versus oysters Analysis of Variance Source Regression Error Total DF 1 166 167 SS 2.01075 2.44516 4.45591 MS 2.01075 0.01473 F 136.51 P 0.000 Same conclusion of significant species effect – F-test is equivalent to t-test for slope parameter (here: oyster). F-test is more general and extends to factors with more than 2 levels … 14 ANOVA with one k-level- factor (k ≥ 3) “One-Way ANOVA” Comparison of means between groups • Linear regression model with k level factor • Assumptions: Normality, Independence, equal variances within groups, Continuous response …. but ANOVA is quite robust and tolerates violations to some extent, if the group sample sizes are similar. 15 For the 3-category species variable Regression Analysis: Cadmium versus CG; ME Predictor Constant CG ME Coef 0.46031 -0.12691 -0.29469 SE Coef 0.02013 0.02609 0.02347 Analysis of Variance Source DF SS Regression 2 2.3174 Residual Error 165 2.1385 Total 167 4.4559 T 22.87 -4.86 -12.56 MS 1.1587 0.0130 P 0.000 0.000 0.000 F 89.40 P 0.000 Overall test of differences between groups 16