Artificial Intelligence and Robotics Methods in Computational Biology: Papers from the AAAI 2013 Workshop
An Evolutionary Search Algorithm to Guide Stochastic Search for
Near-Native Protein Conformations with Multiobjective Analysis
Brian Olson1
Amarda Shehu1,2,3
1
Department of Computer Science,
2
Department of Bioengineering,
3
School of Systems Biology
George Mason University, Fairfax, VA, 22030
Abstract
place of Molecular Dynamics (MD) due to the higher sampling capability of MC.
State-of-the-art protocols handle the high dimensionality of the protein conformational space in two ways. First,
reduced/coarse-grained representations of the protein chain
are employed to lower the number of dimensions. Such representations largely sacrifice side chains, modeling backbone heavy atoms and a designated atom or pseudo-atom
per side chain. Second, the molecular fragment replacement technique is employed to sample new conformations.
Rather than sampling angle values for each of the backbone dihedral angles independently, the technique couples
backbone dihedral angles of consecutive amino acids in a
fragment and samples an entire fragment configuration at a
time from libraries pre-compiled from known protein native
structures (Han and Baker 1996).
Coarse-grained representations and molecular fragment
replacement have greatly advanced de novo structure prediction (Bradley, Misura, and Baker 2005; Hegler et al. 2009;
Shehu 2009; DeBartolo et al. 2010; Shehu and Olson 2010;
Olson, Molloy, and Shehu 2011; Olson et al. 2012b; Xu
and Zhang 2012; Simoncini et al. 2012; Molloy, Saleh,
and Shehu 2013). Recently, this domain-specific expertise has been incorporated in evolutionary search algorithms (EAs) (Olson, De Jong, and Shehu 2013; Olson
and Shehu 2012b; 2013; Saleh, Olson, and Shehu 2012;
2013). EAs have been proposed for protein conformational search before, using either lattice or all-atom representations (Chira, Horvath, and Dumitrescu 2010; Islam, Chetty, and Murshed 2011; Cutello, V et al. 2011;
Garza-Fabre, Toscano-Pulido, and Rodriguez-Tello 2012;
Narzisi, Nicosia, and Stracquadanio 2010). Currently, EAs
that employ lattice or all-atom representations have limited applicability and are not competitive with MC-based
approaches that employ backbone representations. Recent
work has shown that incorporating such representations and
molecular fragment replacement makes even very simple
EAs, such as basin hopping (Olson and Shehu 2011; 2012b;
Olson et al. 2012a; Olson and Shehu 2012a; 2013), or more
powerful population-based EAs (Saleh, Olson, and Shehu
2013; Olson, De Jong, and Shehu 2013) competitive with
MC-based algorithms for de novo structure prediction.
Currently, many stochastic search algorithms are shown
to have high sampling capability. However, inaccuracies
Predicting native conformations of a protein sequence
is known as de novo structure prediction and is a central challenge in computational biology. Most computational protocols employ Monte Carlo sampling. Evolutionary search algorithms have also been proposed to
enhance sampling of near-native conformations. These
approaches bias stochastic search by an energy function, even though current energy functions are known to
be inaccurate and drive sampling to non-native energy
minima. This paper proposes a multiobjective approach
which employs Pareto dominance, rather than total energy, to evaluate a conformation. This multiobjective
approach accounts for the fact that terms in an energy
function are conflicting optimization criteria. Our analysis is conducted on a diverse set of 20 proteins. Results
show that employing Pareto dominance, rather than total energy, to guide stochastic search is more effective at
sampling conformations which are both lower in energy
and near the protein native structure.
Introduction
Millions of protein-encoding sequences extracted from organismal genomes lack any structural or functional characterization (Lee, Redfern, and Orengo 2007). Yet, a detailed structural characterization of the biologically-active
or native state of a protein is key to understanding protein
function and essential in engineering novel proteins, predicting stability, modeling molecular interactions, and designing novel drug compounds (Shehu 2013). Doing so from
only knowledge the protein’s amino-acid sequence, a problem known as de novo structure prediction, is an outstanding challenge in computational biology (Lee, Wu, and Zhang
2009; Shehu 2010; Moult et al. 2011).
Current de novo structure prediction protocols employ
stochastic search guided by an energy function to iterate
over low-energy conformations of a chain of amino acids.
The operating principle is that native conformations are associated with the lowest energies in the energy surface that
underlies the protein conformational space (Dill and Chan
1997). Most protocols use Monte Carlo (MC) sampling in
c 2013, Association for the Advancement of Artificial
Copyright Intelligence (www.aaai.org). All rights reserved.
32
of forward kinematics allows obtaining cartesian coordinates from these angles (Zhang and Kavraki 2002). The only
atoms modeled are the heavy backbone atoms N , Calpha ,
C, O, and a pseudo-atom centered at the side chain of each
amino acid. This representation is the one employed in the
Rosetta de novo structure prediction protocol.
in energy functions are considered primary reasons why
de novo structure prediction remains challenging. Recent
work shows that even state-of-the-art coarse-grained energy
functions, including the Rosetta energy function, have nonnative energy minima that are lower than the one containing the experimentally-known native structure (Shmygelska and Levitt 2009; Das 2011; Molloy, Saleh, and Shehu
2013). This is not surprising, as energy functions, particularly those that interface with coarse-grained representations, are known to be inaccurate due to the process in
which they are obtained. Many terms in them are conflicting. However, this development seems to represent an
impasse in computational structural biology. Recent studies have advocated sacrificing efficiency and doing away
with coarse-grained energy functions (Bowman and Pande
2009), though the presence of inaccuracies is not disputed
even among all-atom energy functions (Verma et al. 2006;
Hornak et al. 2006; Roe et al. 2007).
In this paper we propose to change the framework in
which stochastic search, whether MC- or EA-based, is
guided by an energy function. We propose to treat the different terms or groups of terms in a given energy function
as conflicting optimization criteria. We do so in the context of an EA that combines both local and global search,
which is known as a hybrid or memetic EA. In essence,
the EA evolves a population of conformations over generations. The algorithm employs the coarse-grained representation in the Rosetta protocol, the Rosetta energy functions,
and the molecular fragment replacement technique. Fragment lengths of 9 and 3 amino acids are used.
The proposed algorithm is guided by Pareto dominance
rather than the total potential energy of a conformation. Prior
to adding a conformation to the evolving population, the algorithm decomposes the energy of the conformation into
various terms. The values of these terms are compared to
those of other conformations maintained in an archive. The
conformation is then added to the population based on a
multi-objective analysis detailed below in Methods. The resulting population thus corresponds to the Pareto front of all
of the conformations sampled during the search.
The proposed algorithm is tested on 20 protein sequences
with experimentally-determined native structures. Analysis
of sampled conformations and comparison with the known
native structures show that employing Pareto dominance to
guide stochastic search rather than total energy is more effective at sampling low-energy near-native conformations.
Initial Population
The initial population P0 is obtained by conducting p independent two-stage Metropolis MC trajectories starting at the
fully extended conformation. The first stage of each trajectory consists of 200 moves and uses the score0 Rosetta energy function with a temperature of zero. The second stage
uses the score1 Rosetta energy function with a low temperature close to room temperature. Stage two runs until n
consecutive MC moves have failed, where n is the number
of amino acids. A move in this 0th generation consists of
replacing the configuration of a randomly-selected fragment
of 9 amino acids with a configuration sampled from the fragment configuration library constructed with the latest protocol described in (Leaver-Fay and et al. 2011). The score0
Rosetta energy function consists of only a soft steric repulsion, and its usage in P0 is to obtain a diverse population
of conformations free of steric clashes. The application of
score1 allows formation of secondary structure.
Evolving Population
In each subsequent generation i, the algorithm switches to
employing fragments of length 3, and the population Pi is
obtained as follows. All conformations of the previous population Pi−1 are first duplicated, then subjected to mutation
and projected to a nearby local minimum through a local
search. The mutation consists of replacing a configuration of
a randomly-sampled fragment of length 3. The local search
is a greedy search that terminates when l consecutive replacements fail to lower energy. Analysis in previous work
suggests setting l to the number of amino acids in the target protein sequence (Olson and Shehu 2012a). The energy
function used for the local search is the score3 Rosetta energy function, which corresponds to the full coarse-grained
Rosetta energy function that is a linear combination of
10 different energy terms measuring repulsion, amino-acid
propensities, residue environment, residue pair interactions,
interactions between secondary structure elements, density,
and compactness.
Population Selection
Method
The result of this process is p child conformations which
are not automatically added to population Pi . Instead, they
are compared to an archive that maintains every child conformation sampled in the algorithm. The archive gives a
broad view of conformational space in order to select conformations to add to the population. The comparison is conducted on three groupings of energy terms in the score4
Rosetta energy function. In score4, three additional terms
are added to score3, short-range hydrogen bonding, longrange hydrogen bonding, and Ramachandran. These are organized into three terms, shb, which corresponds to short-
In the proposed EA, a population of conformations evolves
through a series of generations guided by Pareto dominance
(detailed below) rather than the total energy of a conformation. Different fragment lengths and different Rosetta energy
functions are used at various generations.
Molecular Representation
A conformation is represented as a vector of 3n angles,
which are the φ, ψ, ω backbone dihedral angles of each
amino acid in a protein chain of n amino acids. Application
33
In the local search, m is set to the number of amino acids in
the particular protein sequence under consideration.
range hydrogen bonding, lhb, which corresponds to longrange hydrogen bonding, and all-else, which groups together Ramachandran and all other remaining energy terms.
Our analysis indicates that this grouping is most effective
(data not shown). Once the energy of a conformation is split
into these 3 terms, then essentially a conformation can be regarded to have 3 scores. A child conformation is first added
to the archive, and then each conformation in the archive, including the newly added child conformation, is re-evaluated
according to these 3 scores.
Summary Analysis
Table 1 provides details on the 20 protein systems selected
for the analysis here. These vary from 53 to 146 amino acids
in length and have different native folds. The lowest RMSD
to the native structure (also lowest over 5 runs) is shown for
EA in column 5 and compared to that reached by MOEA,
shown in column 6. In 17/20 cases, highlighted in bold,
MOEA reaches lower or comparable (within 0.5Å) lowest
RMSDs. Columns 7-8 show that the % of conformations
with < 5Å from the native structure is also higher in MOEA,
and this difference is dramatic in 3 cases, which are highlighted in bold. In only one case is MOEA outperformed
by EA (protein system 11). Columns 9 − 10 compare the
algorithms in terms of lowest score4 energy value reached.
In 12/20 cases, highlighted in bold, MOEA reaches lower
or comparable (within 2.0kcal/mol) energy values than EA.
In summary, these results suggest that MOEA reaches both
lower-energy and lower-RMSD conformations, thus enhancing sampling of near-native conformations.
Multiobjective Analysis for Selection: Pareto
Dominance
A conformation Ci in the archive is said to dominate another
conformation Cj in the archive when each score of Ci is
lower than the corresponding score in Cj . The Pareto rank of
a conformation is the number of conformations which dominate it. Conformations with a Pareto rank of 0 are said to
be non-dominated and belong to the Pareto front. Conformations in the Pareto front are considered equivalent with
respect to a multiobjective analysis. We note that the Pareto
rank of a conformation in the archive can change over time,
so a conformation that starts in the Pareto front will likely
fall out of the Pareto front over time. This is the reason the
algorithm re-evaluates the entire archive after adding a child
conformation to it.
In addition to child conformations, the best l parent conformations from population Pi−1 are added to population
Pi . This is known as elitism, and its purpose is to preserve
good solutions captured in previous generations. The resulting population is reduced down to the same constant size of
p individuals through truncation selection. For both elitism
and truncation selection, conformations are ranked first by
Pareto rank and then by total energy for conformations with
the same Pareto rank.
Detailed Analysis
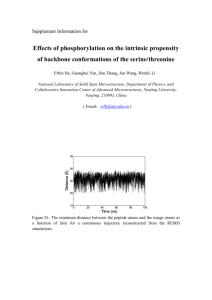
The rest of the analysis provides some more detail on the actual distribution of RMSDs and energies of sampled conformations. The left panel of Fig. 1 compares the distribution of
RMSD values (all conformations sampled by all 5 runs are
combined for each algorithm). The distribution for conformations obtained by MOEA is plotted in a dotted blue line
and is superimposed over that obtained by EA, plotted in
a black line. Three systems are selected to highlight a case
where the MOEA results in significantly more conformations with lower RMSDs to the native structure than the EA
in Fig. 1(a), a case where the distributions are comparable,
shown in Fig. 1(c), and a rare case where EA performs better, shown in Fig. 1(e) (this is the only case where MOEA is
outperformed, as indicated in Table 1).
The right panel of Fig. 1 compares the algorithms in terms
of the energy vs. RMSD distribution of the conformations
they sample (conformations from all 5 runs are combined
for each algorithm). The only three highlighted cases are
those as above. The distribution obtained by the MOEA is
superimposed in blue over that obtained by EA in red. Comparison of these distributions allows making a few observations. First, as before, MOEA reaches lower energy values
than EA, even though it is guided by Pareto analysis rather
than total energy. Second, the energy surfaces sampled are
rich in non-native minima. Only in the case of PDB id 1dtjA
is the energy surface funneled towards the native structure.
In the case PDB id 1aoy, where MOEA performs worse than
EA in terms of distribution of RMSDs, the Pareto analysis
seems to have steered the search towards a minimum that is
7 − 9Å away from the native structure.
Experiments and Results
We compare the proposed algorithm, which we refer to as
MOEA for Multi-objective EA, with an EA that does not
use Pareto rank, but rather only employs total energy to determine whether to add a child conformation to a population
based on truncation selection. Our analysis compares EA to
the MOEA in terms of lowest energies (score4) reached, the
lowest RMSD to the native structure reached, and the entire
distribution of energy vs. RMSD values for sampled conformations. RMSD averages Euclidean distance between corresponding Cα atoms from a given conformation and the
known native structure. Lower values mean better proximity
to the native structure.
Implementation Details
Each algorithm is run 5 times on each of the 20 proteins
employed for our analysis. A fixed budget of 10, 000, 000
energy function evaluations is used, which takes 7−24 hours
of CPU time on a 2.4Ghz Core i7 processor, depending on
protein length. The size of each population is p = 100, and
elitism rate is set to l = 25 for EA and l = 100 for MOEA.
Discussion
Taken together, the results show that guiding search by multiobjective analysis rather than total energy can be more
34
(a) 1dtjA, 76 aas, α/β
(b) 1dtjA, 76 aas, α/β
(c) 1ail, 70 aas, α
(d) 1ail, 70 aas, α/
(e) 1aoy, 66 aas, α/β
(f) 1aoy, 67 aas, α/β
Figure 1: Left: Distribution of RMSDs of MOEA-obtained conformations from known native structure (dotted blue line) are
superimposed over distribution obtained by EA (black line). Right: Distribution of energies vs. RMSDs from native structure
of MOEA-obtained conformations (transparent blue) are superimposed over distribution obtained by EA (red).
35
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
Table 1: Summary of comparison between EA and MOEA on 20 protein sequences.
Native
Fold
min Cα-RMSD (Å) % < 5Å Cα-RMSD
Rosetta score4
PDB Id Length Topology EA
MOEA
EA
MOEA
EA
MOEA
1bq9
53
α/β
3.0
3.4
0.093
0.128
-50.5
-45.8
1dtdB
61
α/β
4.4
5.3
0.006
0.000
-55.0
-74.5
1isuA
62
α/β
6.6
6.4
0.000
0.000
-46.5
-48.4
1c8cA
64
α/β
4.8
3.6
0.001
0.003
-86.4
-98.4
1sap
66
α/β
3.7
3.7
0.015
0.008
-121.4 -120.1
1hz6A
67
α/β
1.9
2.1
13.938
35.418
-130.9 -135.6
1wapA
68
β
6.3
6.4
0.000
0.000
-132.5 -117.5
1fwp
69
α/β
4.3
3.4
0.007
0.107
-84.4
-92.8
1ail
70
α
1.4
1.9
1.747
2.056
-56.1
-67.1
1dtjA
76
α/β
4.2
2.3
0.004
8.174
-82.2
-97.4
1aoy
78
α/β
3.9
3.7
0.368
0.187
-98.1
-102.0
2ci2
83
α/β
3.7
3.9
0.006
0.001
-109.8 -105.7
1cc5
83
α
4.7
4.9
0.001
0.001
-68.6
-67.8
1tig
88
α/β
3.2
2.5
1.095
11.368
-128.0 -151.7
2ezk
93
α
3.4
3.2
0.060
0.493
-100.7
-93.4
1hhp
99
β
8.8
8.6
0.000
0.000
-104.5
-97.3
2hg6
106
α/β
9.3
9.6
0.000
0.000
-102.6
-95.7
3gwl
106
α
5.4
5.8
0.000
0.000
-100.0
-95.3
2h5nD
123
α
6.2
7.5
0.000
0.000
-129.0 -126.6
1aly
146
β
11.2
11.4
0.000
0.000
-81.1
-117.1
effective and enhance sampling of both low-energy and
near-native conformations. This direction seems particularly
promising in the context of inaccurate energy functions and
warrants further investigation in de novo structure prediction. Already researchers in computational biology are investigating multiobjective optimization in the context of protein design (Nivon, Moretti, and Baker 2013). In future work
we will consider different energy functions, different groupings of energy terms, and variations of the Pareto-based
analysis. It is expected that progress in this direction will
not only advance decoy sampling for de novo structure prediction, but it will also provide high-quality decoys for improvements in the process of computational design of protein energy functions.
Chira, C.; Horvath, D.; and Dumitrescu, D. 2010. An Evolutionary Model Based on Hill-Climbing Search Operators
for Protein Structure Prediction. Evolutionary Computation,
Machine Learning and Data Mining in Bioinformatics 38–
49.
Cutello, V; Morelli, G.; Nicosia, G.; Pavone, M.; and Scollo,
G. 2011. On discrete models and immunological algorithms for protein structure prediction. Natural Computing
10(1):91–102.
Das, R. 2011. Four small puzzles that rosetta doesn’t solve.
PLoS ONE 6(5):e20044.
DeBartolo, J.; Hocky, G.; Wilde, M.; Xu, J.; Freed, K. F.; and
Sosnick, T. R. 2010. Protein structure prediction enhanced
with evolutionary diversity: SPEED. Protein Sci. 19(3):520–
534.
Dill, K. A., and Chan, H. S. 1997. From levinthal to pathways to funnels. Nat. Struct. Biol. 4(1):10–19.
Garza-Fabre, M.; Toscano-Pulido, G.; and Rodriguez-Tello,
E. 2012. Locality-based multiobjectivization for the HP
model of protein structure prediction. In GECCO ’12: Proceedings of the fourteenth international conference on Genetic and evolutionary computation conference. ACM Request Permissions.
Han, K. F., and Baker, D. 1996. Global properties of the
mapping between local amino acid sequence and local struc-
Acknowledgment
This work is supported in part by NSF CCF No. 1016995
and NSF IIS CAREER Award No. 1144106.
References
Bowman, G. R., and Pande, V. S. 2009. Simulated tempering yields insight into the low-resolution rosetta scoring
functions. Proteins: Struct. Funct. Bioinf. 74(3):777–788.
Bradley, P.; Misura, K. M.; and Baker, D. 2005. Toward
high-resolution de novo structure prediction for small proteins. Science 309(5742):1868–1871.
36
ture in proteins. Proc. Natl. Acad. Sci. USA 93(12):5814–
5818.
Hegler, J. A.; Laetzer, J.; Shehu, A.; Clementi, C.; and
Wolynes, P. G. 2009. Restriction vs. guidance: fragment assembly and associative memory hamiltonians for
protein structure prediction. Proc. Natl. Acad. Sci. USA
106(36):15302–15307.
Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.;
and Simmerling, C. 2006. Comparison of multiple amber
force fields and development of improved protein backbone
parameters. Proteins: Struct. Funct. Bioinf. 65(3):712–725.
Islam, M. K.; Chetty, M.; and Murshed, M. 2011. Novel local improvement techniques in clustered memetic algorithm
for protein structure prediction. In Evolutionary Computation (CEC), 2011 IEEE Congress on, 1003–1011.
Leaver-Fay, A., and et al. 2011. ROSETTA3: an objectoriented software suite for the simulation and design of
macromolecules. Methods Enzymol 487:545–574.
Lee, D.; Redfern, O.; and Orengo, C. 2007. Predicting protein function from sequence and structure. Nat. Rev. Mol.
Cell Biol. 8(12):995–1005.
Lee, J.; Wu, S.; and Zhang, Y. 2009. Ab initio protein structure prediction. In Rigden, D., ed., Ab Initio Protein Structure Prediction. Springer Science + Business Media B.V.
chapter 1.
Molloy, K.; Saleh, S.; and Shehu, A. 2013. Probabilistic
search and energy guidance for biased decoy sampling in
ab-initio protein structure prediction. IEEE Trans. Comp.
Biol. and Bioinf. in press.
Moult, J.; Fidelis, K.; Kryshtafovych, A.; and Tramontano,
A. 2011. Critical assessment of methods of protein structure
prediction (CASP) round IX. Proteins: Struct. Funct. Bioinf.
Suppl(10):1–5.
Narzisi, G.; Nicosia, G.; and Stracquadanio, G. 2010. Robust Bio-active Peptide Prediction Using Multi-objective
Optimization. In Biosciences (BIOSCIENCESWORLD),
2010 International Conference on, 44–50.
Nivon, L. G.; Moretti, G.; and Baker, D. 2013. A paretooptimal refinement method for protein design scaffolds.
PLoS One 8(4):e59004.
Olson, B., and Shehu, A. 2011. Populating local minima
in the protein conformational space. In IEEE Intl Conf on
Bioinf and Biomed (BIBM), 114–117.
Olson, B., and Shehu, A. 2012a. Efficient basin hopping in
the protein energy surface. In IEEE Intl Conf on Bioinf and
Biomed. in press.
Olson, B., and Shehu, A. 2012b. Evolutionary-inspired
probabilistic search for enhancing sampling of local minima
in the protein energy surface. Proteome Sci. in press.
Olson, B., and Shehu, A. 2013. Rapid sampling of local minima in protein energy surface and effective reduction
through a multi-objective filter. Proteome Sci. in press.
Olson, B.; Hashmi, I.; Molloy, I.; and Shehu, A. 2012a.
Basin hopping as a general and versatile optimization framework for the characterization of biological macromolecules.
Advances in AI J 2012(674832).
Olson, B. S.; Molloy, K.; Hendi, S.-F.; and Shehu, A. 2012b.
Guiding search in the protein conformational space with
structural profiles. J Bioinf and Comp Biol 10(3):1242005.
Olson, S.; De Jong, K. A.; and Shehu, A. 2013. Off-lattice
protein structure prediction with homologous crossover. In
Genet. and Evol. Comput. Conf. (GECCO). in press.
Olson, B.; Molloy, K.; and Shehu, A. 2011. In search of the
protein native state with a probabilistic sampling approach.
J. Bioinf. and Comp. Biol. 9(3):383–398.
Roe, D. R.; Okur, A.; Wickstrom, L.; Hornak, V.; and Simmerling, C. 2007. Secondary structure bias in generalized
born solvent models: Comparison of conformational ensembles and free energy of solvent polarization from explicit and
implicit solvation. J. Phys. Chem. 11(7):1846 –1857.
Saleh, S.; Olson, B.; and Shehu, A. 2012. A populationbased evolutionary algorithm for sampling minima in the
protein energy surface. In He, J.; Shehu, A.; Haspel, N.;
and B., C., eds., Comput Struct Biol Workshop, 48–55.
Saleh, S.; Olson, B.; and Shehu, A. 2013. A populationbased evolutionary search approach to the multiple minima problem in de novo protein structure prediction. BMC
Struct. Biol. in press.
Shehu, A., and Olson, B. 2010. Guiding the search
for native-like protein conformations with an ab-initio treebased exploration. Int. J. Robot. Res. 29(8):1106–11227.
Shehu, A. 2009. An ab-initio tree-based exploration to enhance sampling of low-energy protein conformations. In
Robot: Sci. and Sys., 241–248.
Shehu, A. 2010. Conformational search for the protein native state. In Rangwala, H., and Karypis, G., eds., Protein
Structure Prediction: Method and Algorithms. Fairfax, VA:
Wiley Book Series on Bioinformatics. chapter 21.
Shehu, A. 2013. Probabilistic search and optimization for
protein energy landscapes. In Aluru, S., and Singh, M., eds.,
Handbook of Computational Molecular Biology. Chapman
& Hall/CRC Computer Information Series.
Shmygelska, A., and Levitt, M. 2009. Generalized ensemble
methods for de novo structure prediction. Proc. Natl. Acad.
Sci. USA 106(5):94305–95126.
Simoncini, D.; Berenger, F.; Shrestha, R.; and Zhang, K.
Y. J. 2012. A probabilistic fragment-based protein structure prediction algorithm. PLoS ONE 7(7):e38799.
Verma, A.; Schug, A.; Lee, K. H.; and Wenzel, W. 2006.
Basin hopping simulations for all-atom protein folding. J.
Chem. Phys. 124(4):044515.
Xu, D., and Zhang, Y. 2012. Ab initio protein structure assembly using continuous structure fragments and optimized
knowledge-based force field. Proteins: Struct. Funct. Bioinf.
80(7):1715–1735.
Zhang, M., and Kavraki, L. E. 2002. A new method for fast
and accurate derivation of molecular conformations. Chem.
Inf. Comput. Sci. 42(1):64–70.
37